For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RFGFLFNEVVTGLRRNVTMTVAMIITTAIAIGLFGGGLLVISLAKNSKAIYLDRVETQIFLTEDLSATDT TCSSDICKGLRDEIEKRSDIKSVRFVNREDAYADAGRRLPQFQDLMKDVSKDAFPASFIVKLKDPEKHAD FDEAFVGKPGVRGVLNQKDLIDRLFAVLDSLRDAAFMIALIQAVGAVLLIANMVQVAAYTRRTEVGIMRL VGATRWYTQLPFLLEAVIAALAGVALAVIGLIIARVTILNGALQQFIQANLVAPITYGDVFLAAIQMAAL GILLAGVTAYVTLRLYVRR*
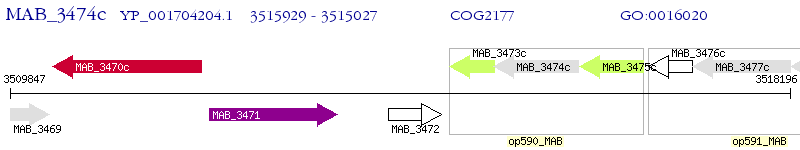
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3474c | - | - | 100% (300) | cell division protein FtsX-like protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3128c | ftsX | 1e-112 | 65.00% (300) | putative cell division protein FTSX (septation |
| M. gilvum PYR-GCK | Mflv_4447 | - | 1e-114 | 67.44% (301) | hypothetical protein Mflv_4447 |
| M. tuberculosis H37Rv | Rv3101c | ftsX | 1e-112 | 65.00% (300) | putative cell division protein FTSX (septation |
| M. leprae Br4923 | MLBr_00670 | ftsX | 1e-115 | 66.00% (300) | hypothetical protein MLBr_00670 |
| M. marinum M | MMAR_1531 | ftsX | 1e-117 | 69.33% (300) | cell division protein FtsX |
| M. avium 104 | MAV_4002 | - | 1e-115 | 67.33% (300) | efflux ABC transporter, permease protein |
| M. smegmatis MC2 155 | MSMEG_2090 | - | 1e-113 | 67.44% (301) | putative cell division protein FtsX |
| M. thermoresistible (build 8) | TH_2154 | ftsX | 1e-113 | 67.33% (300) | PUTATIVE CELL DIVISION PROTEIN FTSX (SEPTATION |
| M. ulcerans Agy99 | MUL_2408 | ftsX | 1e-116 | 69.00% (300) | cell division protein FtsX |
| M. vanbaalenii PYR-1 | Mvan_1915 | - | 1e-113 | 66.11% (301) | hypothetical protein Mvan_1915 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00670|M.leprae_Br4923 MRFGFLLNEVVTGLRRNVTMTIAMILTTAISIGLFGGGLLVVRLADNSRS
MAV_4002|M.avium_104 MRFGFLLNEVLTGLRRNVTMTAAMILTTAISIGLFGGGLLVVRLADNSRE
Mb3128c|M.bovis_AF2122/97 MRFGFLLNEVLTGFRRNVTMTIAMILTTAISVGLFGGGMLVVRLADSSRA
Rv3101c|M.tuberculosis_H37Rv MRFGFLLNEVLTGFRRNVTMTIAMILTTAISVGLFGGGMLVVRLADSSRA
MMAR_1531|M.marinum_M MRFGFLLNEVLTGLRRNVTMTVAMILTTAISIGLFGGGLLVVRLAENSRA
MUL_2408|M.ulcerans_Agy99 MRFGFLLNEVRTGLRRNVTMTVAMILTTAISIGLFGGGLLVVRLAENSRA
Mflv_4447|M.gilvum_PYR-GCK MRFGFLVNEVLTGLRRNVTMTVAMILTTAISIGLFGGGMLVVRLADQSRA
Mvan_1915|M.vanbaalenii_PYR-1 MRFGFLVNEVLTGLRRNVTMTVAMILTTAISIGLFGGGMLIVRLADQSRA
MSMEG_2090|M.smegmatis_MC2_155 MRFGFLINEVLTGLRRNVTMTVAMVLTTAISIGLFGGGLLVVRLADQSRD
TH_2154|M.thermoresistible__bu VRFGFLLNEVLTGFRRNVTMTVAMILTTAISIGLFGGGLLVLRLADHSKN
MAB_3474c|M.abscessus_ATCC_199 MRFGFLFNEVVTGLRRNVTMTVAMIITTAIAIGLFGGGLLVISLAKNSKA
:*****.*** **:******* **::****::******:*:: **. *:
MLBr_00670|M.leprae_Br4923 IYLDRVETQVFLTDDISANDLTCNTNLCKALRGKIEARDDVKSLRFLNRQ
MAV_4002|M.avium_104 IYLDRVETQVFLTDDISANDATCGSAPCKALRDKIDARQDVKSVRFVNRQ
Mb3128c|M.bovis_AF2122/97 IYLDRVESQVFLTEDVSANDSSCDTTACKALREKIETRSDVKAVRFLNRQ
Rv3101c|M.tuberculosis_H37Rv IYLDRVESQVFLTEDVSANDSSCDTTACKALREKIETRSDVKAVRFLNRQ
MMAR_1531|M.marinum_M IYLDRVETQVFLTDDVSANDPNCDSDPCKALREKIEKRKDVKAVRFLNRA
MUL_2408|M.ulcerans_Agy99 IYLDRVETQVFLTDDVSANDPNCDSDPCKALREKIGKRKDVKAVRFLNRA
Mflv_4447|M.gilvum_PYR-GCK IYLDRVESQVFLTPDASADDPTCDNDPCKALRQTIEDRDDVRSVRFLNRD
Mvan_1915|M.vanbaalenii_PYR-1 IYLDRVESQVFLTNDVSANDPSCDADPCKALRQTIEERDDVRSVRFLNRD
MSMEG_2090|M.smegmatis_MC2_155 IYLDRVESQVFLTDDISANDPTCDSDPCKALYRKIDDRDDVRSLRFLNRE
TH_2154|M.thermoresistible__bu IYLDRVESQVFLTNDVSANDPTCDADPCQALREQIEARDDVRSVRFLNRD
MAB_3474c|M.abscessus_ATCC_199 IYLDRVETQIFLTEDLSATDTTCSSDICKGLRDEIEKRSDIKSVRFVNRE
*******:*:*** * ** * .*. *:.* * *.*::::**:**
MLBr_00670|M.leprae_Br4923 DAYDDAIRKFPQYRDVAG---KDSFPASFIIKLANPVQHKEFDAATQGQP
MAV_4002|M.avium_104 DAYNDAIKKFPEFKDVAG---KDSFPASFIVKLNNPEQHAEFDAAMQGQP
Mb3128c|M.bovis_AF2122/97 QAYDDAIRKFPQFKDVAG---KDSFPASFIVKLENPEQHKDFDTAMKGQP
Rv3101c|M.tuberculosis_H37Rv QAYDDAIRKFPQFKDVAG---KDSFPASFIVKLENPEQHKDFDTAMKGQP
MMAR_1531|M.marinum_M QAYDDAIRKFPQYKDVAG---KDSFPASFIVKLENPEQHKDFDSAMQGQP
MUL_2408|M.ulcerans_Agy99 QAYDDAIRKFPQYKDVAG---KDSFPASFIVKLENPEQHKDFDSAMQGQP
Mflv_4447|M.gilvum_PYR-GCK EAYEDAIRKFPQYKDVAG---RDAFPASFVVKLDDPEQHQNFDEAMVGQP
Mvan_1915|M.vanbaalenii_PYR-1 EAYDDAIRKFPQYKDVAG---RDAFPASFVVKLNDPEQHQAFDDAVRGQP
MSMEG_2090|M.smegmatis_MC2_155 QAYDDAIKKFPQYKDVAG---KDAFPASFVVKLVDPENHEEFDQAMVGQP
TH_2154|M.thermoresistible__bu AAYDDAISKFPQYKDVAG---RDAFPASFIVKLHDPEQHQQFDEAMVGQP
MAB_3474c|M.abscessus_ATCC_199 DAYADAGRRLPQFQDLMKDVSKDAFPASFIVKLKDPEKHADFDEAFVGKP
** ** ::*:::*: :*:*****::** :* :* ** * *:*
MLBr_00670|M.leprae_Br4923 GVLSVLNQKELIDRLFAVLDGLSDVAFVIALVQAIGAILLIANMVQVAAY
MAV_4002|M.avium_104 GVRSLLNEKDLIDRLFAVLDGLRNAAFAVALVQAVGAILLIANMVQVAAH
Mb3128c|M.bovis_AF2122/97 GVLDVLNQKELIDRLFAVLDGLSSAAFAVALVQAIGAILLIANMVQVAAY
Rv3101c|M.tuberculosis_H37Rv GVLDVLNQKELIDRLFAVLDGLSNAAFAVALVQAIGAILLIANMVQVAAY
MMAR_1531|M.marinum_M GVLSVLNQKDLIDRLFAVLDGLSNAAFAVALVQAVGAILLIANMVQVAAY
MUL_2408|M.ulcerans_Agy99 GVLSVLNQKDLIDRLFAVLDGLSNAAFAVALVQAVGAILLIANMVQVAAY
Mflv_4447|M.gilvum_PYR-GCK GVQSVLNQKDLIDRLFAVLDGISNVAFAVALVQAIGAVLLIANMVQVAAY
Mvan_1915|M.vanbaalenii_PYR-1 GVLNVLNQKDLIDRLFAVLDGISNAAFAVALVQAIGAVLLIANMVQVAAY
MSMEG_2090|M.smegmatis_MC2_155 GVLNVLNQKDLIDRLFAVLDGISSAAFAVALVQAIGAVLLIANMVQVAAY
TH_2154|M.thermoresistible__bu GVLNVLNQKELIDRLFAVLDGLSNAAFAVALVQAIGAILLIANMVQVAAY
MAB_3474c|M.abscessus_ATCC_199 GVRGVLNQKDLIDRLFAVLDSLRDAAFMIALIQAVGAVLLIANMVQVAAY
** .:**:*:**********.: ..** :**:**:**:***********:
MLBr_00670|M.leprae_Br4923 TRRTEIGIMRLVGASRWYTQLPFLLEAMVAATVGAVIAIVGLLVARAMFL
MAV_4002|M.avium_104 TRRTEIGIMRLVGASRWYTQLPFLVEAMLAAAIGVAIAVVGLILVRAWFL
Mb3128c|M.bovis_AF2122/97 TRRTEIGIMRLVGASRWYTQLPFLVEAMLAATMGVGIAVAGLMVVRALFL
Rv3101c|M.tuberculosis_H37Rv TRRTEIGIMRLVGASRWYTQLPFLVEAMLAATMGVGIAVAGLMVVRALFL
MMAR_1531|M.marinum_M TRRTEIGIMRLVGASRWYTQLPFLMEAMLAATLGVVIAIGGLIFVRAMFL
MUL_2408|M.ulcerans_Agy99 TRRTEIGIMRLVGASRWYTQLPFLMEAMLAATLGVVIAIGGLIFVRAMFL
Mflv_4447|M.gilvum_PYR-GCK TRRTEIGIMRLVGATRWYTQLPFLVEAMLAAFIGVVLAIVGLILVRALFL
Mvan_1915|M.vanbaalenii_PYR-1 TRRTEIGIMRSVGATRWYTQLPFLLEAMLAALIGVVIAIVGLILVRALFL
MSMEG_2090|M.smegmatis_MC2_155 TRRTEIGIMRLVGATRWYTQLPFLVEAMLAALIGVILAIVGLIAVRAMFL
TH_2154|M.thermoresistible__bu TRRTEVGIMRLVGASRWYTQLPFLVEAMLAALIGVVIAIIGLIVVRSLFL
MAB_3474c|M.abscessus_ATCC_199 TRRTEVGIMRLVGATRWYTQLPFLLEAVIAALAGVALAVIGLIIARVTIL
*****:**** ***:*********:**::** *. :*: **: .* :*
MLBr_00670|M.leprae_Br4923 NNALNQFYQANLIARVDYADVLYVS-PWLLLLGVALAALTGYATLRIYVR
MAV_4002|M.avium_104 DNALSQFYQAHLIAKVDYADILYIS-PWLFVLGVSIAGLTAYATLRLYIR
Mb3128c|M.bovis_AF2122/97 ENALNQFYQANLIAKVDYADILFIT-PWLLLLGVAMSGLTAYLTLRLYVR
Rv3101c|M.tuberculosis_H37Rv ENALNQFYQANLIAKVDYADILFIT-PWLLLLGVAMSGLTAYLTLRLYVR
MMAR_1531|M.marinum_M ENALNQFYQANLIARIDYADIFFIS-PALLALGVAMAGATAYVTLRLYVR
MUL_2408|M.ulcerans_Agy99 ENALNQFYQANLIARIDYADIFFIS-PALLALGVAMAGATAYVTLRLYVR
Mflv_4447|M.gilvum_PYR-GCK EKALDQFYQANLIAQVDYADILYYSAPWMLFLGLAMSGITAYVTLRLYVR
Mvan_1915|M.vanbaalenii_PYR-1 EKALDQFYQANLIAKVDYADILYYSAPWMLILGLAMSGITSYVTLRLYVR
MSMEG_2090|M.smegmatis_MC2_155 EKALDQFYQANLIAKVDYADILYYSAPWMLFLGLAMSGVTAYVTLRLYVR
TH_2154|M.thermoresistible__bu EKALDQFYQANLIARIDYLDIVYIS-PILLVVGVAMAGITAYVTLRLYVR
MAB_3474c|M.abscessus_ATCC_199 NGALQQFIQANLVAPITYGDVFLAA-IQMAALGILLAGVTAYVTLRLYVR
: **.** **:*:* : * *:. : : :*: ::. *.* ***:*:*
MLBr_00670|M.leprae_Br4923 R
MAV_4002|M.avium_104 R
Mb3128c|M.bovis_AF2122/97 R
Rv3101c|M.tuberculosis_H37Rv R
MMAR_1531|M.marinum_M R
MUL_2408|M.ulcerans_Agy99 R
Mflv_4447|M.gilvum_PYR-GCK R
Mvan_1915|M.vanbaalenii_PYR-1 R
MSMEG_2090|M.smegmatis_MC2_155 R
TH_2154|M.thermoresistible__bu R
MAB_3474c|M.abscessus_ATCC_199 R
*