For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TNIAVLIKQVPDTWSERKLTDGDFTLDREAADAVLDEINERAVEEALLIKEREGGDSVVTVVSAGPEKAT EAIRKALSMGADKAVHLLDPGLHGSDAIQTGWALARALGQVEGVELVIAGNEATDGRSGAIPAIIAEYLG LPQLTHLRKLTVEGGVVKGERETDEGVFEVEATLPAIVSVTEKINEPRFPSFKGIMAAKKKEVAVLTLAE IGVEADEVGVANAGSTVLSSTPKAAKTAGEKITDEGEGGTKVAEFLVAQKII*
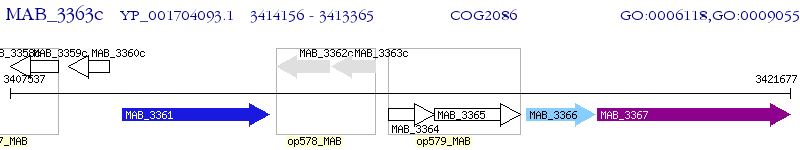
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3363c | - | - | 100% (263) | electron transfer flavoprotein subunit beta FixA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3055c | fixA | 1e-123 | 81.95% (266) | electron transfer flavoprotein subunit beta |
| M. gilvum PYR-GCK | Mflv_4259 | - | 1e-126 | 85.93% (263) | electron transfer flavoprotein beta-subunit |
| M. tuberculosis H37Rv | Rv3029c | fixA | 1e-123 | 81.95% (266) | electron transfer flavoprotein subunit beta |
| M. leprae Br4923 | MLBr_01712 | eftB | 1e-120 | 81.20% (266) | electron transfer flavoprotein beta subunit |
| M. marinum M | MMAR_1684 | fixA | 1e-124 | 84.03% (263) | electron transfer flavoprotein (beta-subunit) FixA |
| M. avium 104 | MAV_3876 | - | 1e-124 | 83.27% (263) | electron transfer protein, beta subunit |
| M. smegmatis MC2 155 | MSMEG_2351 | etfB | 1e-124 | 84.03% (263) | electron transfer flavoprotein, beta subunit |
| M. thermoresistible (build 8) | TH_2194 | fixA | 1e-120 | 81.75% (263) | PROBABLE ELECTRON TRANSFER FLAVOPROTEIN (BETA-SUBUNIT) FIXA |
| M. ulcerans Agy99 | MUL_1922 | fixA | 1e-123 | 83.65% (263) | electron transfer flavoprotein (beta-subunit) FixA |
| M. vanbaalenii PYR-1 | Mvan_2099 | - | 1e-125 | 84.79% (263) | electron transfer flavoprotein beta-subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2351|M.smegmatis_MC2_155 MTNIVVLIKQVPDTWSERKLSDGDFTLDREAADAVLDEINERAVEEALLI
TH_2194|M.thermoresistible__bu MTNIVVLIKQVPDTWSERKLRDDDFTLDREAADAVLDEINERAVEEALLI
Mflv_4259|M.gilvum_PYR-GCK MTNIVVLIKQVPDTWSERKLSEGDWTLDREAADAVLDEINERAVEEALLI
Mvan_2099|M.vanbaalenii_PYR-1 MTNIVVLIKQVPDTWSERKLSDGDYTLDREAADAVLDEINERAVEEALLI
Mb3055c|M.bovis_AF2122/97 MTNIVVLIKQVPDTWSERKLTDGDFTLDREAADAVLDEINERAVEEALQI
Rv3029c|M.tuberculosis_H37Rv MTNIVVLIKQVPDTWSERKLTDGDFTLDREAADAVLDEINERAVEEALQI
MLBr_01712|M.leprae_Br4923 MTNIVVLIKQVPDTWSERKLTDGDFTLDREAADAVLDEINERAVEEALQI
MMAR_1684|M.marinum_M MTNIVVLIKQVPDTWSERKLTEGDWTLDREAADAVLDEINERAVEEALQI
MUL_1922|M.ulcerans_Agy99 MTNIVVLIKQVPDTWSERKLTEGDWTLDREAADAVLDEINERAVEEALQI
MAV_3876|M.avium_104 MTNIVVLIKQVPDTWSERKLTDGDWTLDREAADAVLDEINERAVEEALQI
MAB_3363c|M.abscessus_ATCC_199 MTNIAVLIKQVPDTWSERKLTDGDFTLDREAADAVLDEINERAVEEALLI
****.*************** :.*:*********************** *
MSMEG_2351|M.smegmatis_MC2_155 KEREGGD---STVTVLTAGPERATEAIRKALSMGADKAVHVKDDGLHGSD
TH_2194|M.thermoresistible__bu KEREGGE---GTVTVLTAGPERATEAIRKALSMGADKAVHLLDDGMKGSD
Mflv_4259|M.gilvum_PYR-GCK KEREGGD---STVTVLTAGPERATEAIRKALSMGADKAVHLLDDGLHGSD
Mvan_2099|M.vanbaalenii_PYR-1 KEREGGD---STVTVLTAGPERATEAIRKALSMGADKAVHLVDEGMHGSD
Mb3055c|M.bovis_AF2122/97 REKEAADGIEGSVTVLTAGPERATEAIRKALSMGADKAVHLKDDGMHGSD
Rv3029c|M.tuberculosis_H37Rv REKEAADGIEGSVTVLTAGPERATEAIRKALSMGADKAVHLKDDGMHGSD
MLBr_01712|M.leprae_Br4923 REKEAVDGIKGSVTVLTAGPERATEAIRKALSMGADKAVHLKDDGMHGSD
MMAR_1684|M.marinum_M REREGGE---GSVTVLTAGPERATEAIRKALSMGADKAVHLKDDGMHGSD
MUL_1922|M.ulcerans_Agy99 REREGGE---GSVTVLTAGPERATEAIRKALSMGADKAVHLKDDGMRGSD
MAV_3876|M.avium_104 REREGGE---GSVTVLTAGPERATEAIRKALSMGADKAVHLKDDGMHGSD
MAB_3363c|M.abscessus_ATCC_199 KEREGGD---SVVTVVSAGPEKATEAIRKALSMGADKAVHLLDPGLHGSD
:*:*. : . ***::****:******************: * *::***
MSMEG_2351|M.smegmatis_MC2_155 LIQTAWTLARALGTIEGTELVIAGNEATDGVGGAVPAVIAEYLGLPQLTH
TH_2194|M.thermoresistible__bu MVQTAWALARALGTIEGTELVIAGNESTDGVGAAVPAIVAEYLGLPQLTH
Mflv_4259|M.gilvum_PYR-GCK MVQTGWALARALGTIEGTELVIAGNEATDGTGGAVPAIIAEYLGLPQLTH
Mvan_2099|M.vanbaalenii_PYR-1 MVQTGWALARALGTIEGTELVIAGNEATDGTGGAVPAIIAEYLGLPQLTH
Mb3055c|M.bovis_AF2122/97 VIQTGWALARALGTIEGTELVIAGNESTDGVGGAVPAIIAEYLGLPQLTH
Rv3029c|M.tuberculosis_H37Rv VIQTGWALARALGTIEGTELVIAGNESTDGVGGAVPAIIAEYLGLPQLTH
MLBr_01712|M.leprae_Br4923 VIQTGWALARALGTIEGTELVIAGNESTDGVGGVVPAIIAEYLGLPQLTH
MMAR_1684|M.marinum_M VIQTGWALARALGAIEGTELVIAGNESTDGVGGAVPAIIAEYLGLPQLTH
MUL_1922|M.ulcerans_Agy99 VIQTGWALARALGAIEGTELVIAGNESTDGVGGAVPAIIAEYLGLPQLTH
MAV_3876|M.avium_104 VVQTGWALARALGTIEGTELVIAGNESTDGTGGAVPAIIAEYLGLPQLTH
MAB_3363c|M.abscessus_ATCC_199 AIQTGWALARALGQVEGVELVIAGNEATDGRSGAIPAIIAEYLGLPQLTH
:**.*:****** :**.********:*** ...:**::***********
MSMEG_2351|M.smegmatis_MC2_155 VRKLNVENGKVTAERETDEGVFTLEASLPAVVSVNEKINEPRFPSFKGIM
TH_2194|M.thermoresistible__bu VRKLTVENGKVIAERETDDGVFTLEASLPAVVSVNEKINEPRFPSFKGIM
Mflv_4259|M.gilvum_PYR-GCK VRKLSVENGKVTGERETDDGLFTLEASLPAVVSVNEKINEPRFPSFKGIM
Mvan_2099|M.vanbaalenii_PYR-1 VRKLSVENGKVTAERETDDGVFTLEASLPAVVSVNEKINEPRFPSFKGIM
Mb3055c|M.bovis_AF2122/97 LRKVSIEGGKITGERETDEGVFTLEATLPAVISVNEKINEPRFPSFKGIM
Rv3029c|M.tuberculosis_H37Rv LRKVSIEGGKITGERETDEGVFTLEATLPAVISVNEKINEPRFPSFKGIM
MLBr_01712|M.leprae_Br4923 LRTISVEGGKITGERETDEGVFTLEAVLPAVVSVNEKINEPRFPSFKGIM
MMAR_1684|M.marinum_M VRKLSVEGGKVTGERETDEGLFTLEASLPAVVSVNEKINEPRFPSFKGIM
MUL_1922|M.ulcerans_Agy99 VRKLSVEGGKVTGERETDEGLFTLEASLPAVVSVNEKINEPRFPSFKGIM
MAV_3876|M.avium_104 LRKLSIENGKVTGERETDDGVFNLEATLPAVVSVTEKINEPRFPSFKGIM
MAB_3363c|M.abscessus_ATCC_199 LRKLTVEGGVVKGERETDEGVFEVEATLPAIVSVTEKINEPRFPSFKGIM
:*.:.:*.* : .*****:*:* :** ***::**.***************
MSMEG_2351|M.smegmatis_MC2_155 AAKKKEVTVLSLAEIGVEPDEVGVANAGSKVLASTPKPPKTAGEKVTDEG
TH_2194|M.thermoresistible__bu AAKKKEVTTLTLADIGVEPDEVGVENAGSRVLSATPKPPKTAGEKITDEG
Mflv_4259|M.gilvum_PYR-GCK AAKKKEVTTLTLAEIGVEADEVGVANAGSKVLSSTPKPPKTAGEKVTDEG
Mvan_2099|M.vanbaalenii_PYR-1 AAKKKEVTKLTLAEIGVESDEVGVANAGSKVLSSTPKPPKTAGEKVTDEG
Mb3055c|M.bovis_AF2122/97 AAKKKEVTVLTLAEIGVESDEVGLANAGSTVLASTPKPAKTAGEKVTDEG
Rv3029c|M.tuberculosis_H37Rv AAKKKEVTVLTLAEIGVESDEVGLANAGSTVLASTPKPAKTAGEKVTDEG
MLBr_01712|M.leprae_Br4923 AAKKKEVTVLTLAEIGVEVDEVGLANAGSKVLVSTPKPAKTAGEKITDEG
MMAR_1684|M.marinum_M AAKKKEVTVLTLAEIGVEADEVGLANAGSTVLSSTPKPAKTAGEKVTDEG
MUL_1922|M.ulcerans_Agy99 AAKKKEVTVLTLAEIGVEADEVGLANAGSTVLSSTPKPAKTAGEKVTDEG
MAV_3876|M.avium_104 AAKKKEVTVLTLAEIGVEPDEVGLANAGSSVLSSTPKPPKTAGEKVTDEG
MAB_3363c|M.abscessus_ATCC_199 AAKKKEVAVLTLAEIGVEADEVGVANAGSTVLSSTPKAAKTAGEKITDEG
*******: *:**:**** ****: **** ** :***..******:****
MSMEG_2351|M.smegmatis_MC2_155 DGGTKIAEYLVAQKLI
TH_2194|M.thermoresistible__bu DGGNKIAEFLVAQKLI
Mflv_4259|M.gilvum_PYR-GCK EGGKKVAEYLVAQKII
Mvan_2099|M.vanbaalenii_PYR-1 DGGTKIAEYLVAQKII
Mb3055c|M.bovis_AF2122/97 EGGNQIVQYLVAQKII
Rv3029c|M.tuberculosis_H37Rv EGGNQIVQYLVAQKII
MLBr_01712|M.leprae_Br4923 DGGNQIVQYLVAQKII
MMAR_1684|M.marinum_M EGGNQIVQYLVAQKII
MUL_1922|M.ulcerans_Agy99 EGGNQIVQYLVAQKII
MAV_3876|M.avium_104 EGGNQIAQYLVGQKII
MAB_3363c|M.abscessus_ATCC_199 EGGTKVAEFLVAQKII
:**.::.::**.**:*