For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
APNVLSVSMWAKGTGLGSWPGQQARDATEIVVAELPLPHLVELPARGVGADLIGRAGALLVDIAIDVSPT GYRTTARPGAVVRRARGFLDEDMDALEEEWERGGHRGSERAVKVQGPGPVTLAAELELPNGHRAITDPGA VRDVAASLAEGLAQHRATLSRRLGVPVMVQIDEPSLPAALDGRLSGTSMFSPVHPVDVPTATALLDQCAE AVGGELLVHCCASDPPWELLREANIRAIAIDVAHLADAHALDGLASVLDTGKAVVLGVIPAQAPSTPISS DVLATRTAALADKLGFGRAMLRDRVGVSAGCGLAGTDLQWARKATELTRKVSDLIDADPDAL*
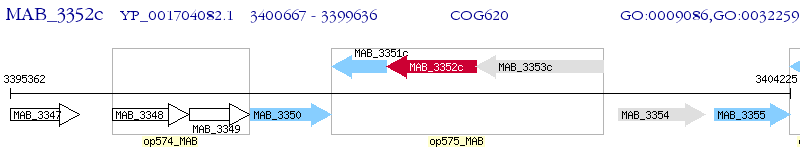
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3352c | - | - | 100% (343) | putative methionine synthase, vitamin-B12 independent |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3040c | - | 1e-106 | 57.69% (338) | hypothetical protein Mb3040c |
| M. gilvum PYR-GCK | Mflv_4252 | - | 1e-113 | 59.52% (336) | methionine synthase, vitamin-B12 independent |
| M. tuberculosis H37Rv | Rv3015c | - | 1e-106 | 57.69% (338) | hypothetical protein Rv3015c |
| M. leprae Br4923 | MLBr_01706 | - | 1e-102 | 55.62% (338) | hypothetical protein MLBr_01706 |
| M. marinum M | MMAR_1692 | - | 1e-109 | 58.58% (338) | hypothetical protein MMAR_1692 |
| M. avium 104 | MAV_3869 | - | 1e-108 | 58.21% (335) | methionine synthase, vitamin-B12 independent, putative |
| M. smegmatis MC2 155 | MSMEG_2359 | - | 1e-106 | 57.57% (337) | methionine synthase, vitamin-B12 independent |
| M. thermoresistible (build 8) | TH_1187 | - | 1e-105 | 57.31% (335) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1930 | - | 1e-107 | 57.69% (338) | hypothetical protein MUL_1930 |
| M. vanbaalenii PYR-1 | Mvan_2108 | - | 1e-109 | 58.46% (337) | methionine synthase, vitamin-B12 independent |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01706|M.leprae_Br4923 -------MSVFA----TASGVGSWPGSSPYPAAKVVVGELAGALAHIVEL
MAV_3869|M.avium_104 -------MSAFARPFATASGVGSWPGTVAREAAEVVVGELAGALAHIVEL
Mb3040c|M.bovis_AF2122/97 -------MSVFA----TATGIGSWPGTAAREAAQVVVGELAGALAYLTEL
Rv3015c|M.tuberculosis_H37Rv -------MSVFA----TATGIGSWPGTAAREAAQVVVGELAGALAYLTEL
MMAR_1692|M.marinum_M -------MSVFA----TATGVGSWPGILARPAAEVVVGELADALAYLVEL
MUL_1930|M.ulcerans_Agy99 -------MSVFA----TATGVGSWPGILARPAAEVVVGELADALAYLVEL
Mflv_4252|M.gilvum_PYR-GCK -------MSVFA----TATGVGSWPGTSAKEAAEVVVGELH-RLPHLVEL
Mvan_2108|M.vanbaalenii_PYR-1 -------MSVFA----AATGIGSWPGTSAREAAEVVIGELH-RLPHLVEL
MSMEG_2359|M.smegmatis_MC2_155 -------MSIFA----TATGVGSWPGTTARQAAEIVVGELH-TLSHLVEL
TH_1187|M.thermoresistible__bu -------VSVFA----VATGVGSWPGTSAREAAQVVVGELA-GLPHLVEL
MAB_3352c|M.abscessus_ATCC_199 MAPNVLSVSMWA----KGTGLGSWPGQQARDATEIVVAELP--LPHLVEL
:* :* .:*:***** . *:::*:.** *.::.**
MLBr_01706|M.leprae_Br4923 PARGVGADMLGRAGALLIDVAIDTVPRGYRIAARPGAVTRRAFSLLDEDM
MAV_3869|M.avium_104 PARGVGADMLGRAGALLVDVAIDTVPRGYRLVARPGAVTRRAISLLDEDM
Mb3040c|M.bovis_AF2122/97 PARGVGADMLGRAGGLLVDVAIDTVPRGYRIAARPGAVTRRAASLLDEDM
Rv3015c|M.tuberculosis_H37Rv PARGVGADMLGRAGGLLVDVAIDTVPRGYRIAARPGAVTRRAASLLDEDM
MMAR_1692|M.marinum_M PARGVGADMLGRAGALLVDLALDTVPRGYRIVSRPGAVTRRAISMLNEDM
MUL_1930|M.ulcerans_Agy99 PARGVGADMLGRAGALLVDLALDTVPRGYRIVPRPGAVTRRAISMLNEDM
Mflv_4252|M.gilvum_PYR-GCK PARGLGADMIGRAGGLLVDIWIDTVPRGYRIASGRSSVTRRAVSLLEEDL
Mvan_2108|M.vanbaalenii_PYR-1 PARGVGADMIGRAGALLVDISIDTVPRGYRIARGRSGAMRRAVSLLEEDL
MSMEG_2359|M.smegmatis_MC2_155 PARGIGADLIGRAGALLVDIGIDTVPRGYRIAPGRSSALRRAVSLLGEDL
TH_1187|M.thermoresistible__bu PERGIGADVIGRAAALLIDIGMDTVPRGYRVASRRTRVVRRAVSLLDEDL
MAB_3352c|M.abscessus_ATCC_199 PARGVGADLIGRAGALLVDIAIDVSPTGYRTTARPGAVVRRARGFLDEDM
* **:***::***..**:*: :*. * *** . . *** .:* **:
MLBr_01706|M.leprae_Br4923 DAFEAAWEMAGLRGRGRVVKVQAPGPITLAAGLELANGHRAITDSCAVRD
MAV_3869|M.avium_104 DALEEAWEAAGLRNDGRVVKVQAPGPVTLAAEIELANGHRAITDPGALRD
Mb3040c|M.bovis_AF2122/97 DALEEAWETAGLRGCGRAVKVQAPGPVTLVAGLELANGHRAITDPGAVRD
Rv3015c|M.tuberculosis_H37Rv DALEEAWETAGLRGCGRAVKVQAPGPVTLVAGLELANGHRAITDPGAVRD
MMAR_1692|M.marinum_M DALEEAWETAGLRGGGRVVKVQAPGPITLAAGVELSNGHRAITDPGAVRD
MUL_1930|M.ulcerans_Agy99 DALEEAWETAGLRGGGRVVKVQAPGPITLAAGVELSNGHRAIIDPGAVRD
Mflv_4252|M.gilvum_PYR-GCK DALEEAWEKAGLRGGGRAVKVQAPGPVTLAAQLELPGGHRAITDPGAVRD
Mvan_2108|M.vanbaalenii_PYR-1 DALEEAWEKAGLRGGERAVKVQAPGPVTLAAQLELPGGHRAITDQGAVRD
MSMEG_2359|M.smegmatis_MC2_155 DALEEAWEKAGLRGSGRPVKLQAPGPVTLAAHLELPGGHRAITDPGALRD
TH_1187|M.thermoresistible__bu DALEEAWERAGLRGTSRSVKVQAPGPITLAAEVELANGHRAVTDTGALRD
MAB_3352c|M.abscessus_ATCC_199 DALEEEWERGGHRGSERAVKVQGPGPVTLAAELELPNGHRAITDPGAVRD
**:* ** .* *. * **:*.***:**.* :**..****: * *:**
MLBr_01706|M.leprae_Br4923 LAESLAEGVAAHRAALARRLDTQVVVQFDEVSLPAALGGLLTGVTAFSPV
MAV_3869|M.avium_104 LAASLGEGVAAHRARLSRRLETPVVVQFDEPSLPAALGGGLPGVTALSPV
Mb3040c|M.bovis_AF2122/97 LAASLAEGVAAHRAALARRLDTPVVVQFDEPSLPAALGGRLTGVTALSPV
Rv3015c|M.tuberculosis_H37Rv LAASLAEGVAAHRAALARRLDTPVVVQFDEPSLPAALGGRLTGVTALSPV
MMAR_1692|M.marinum_M LAASLGEGVAAHRAELARRLDTPVVVQFDEPTLPAALEGRLTGVTSLAPV
MUL_1930|M.ulcerans_Agy99 LAASLGEGVAAHRAELARRLDTPVVVQFDEPTLPAALEGRLTGVTSLAPV
Mflv_4252|M.gilvum_PYR-GCK LTASLAEGVAAHRAALARRLESPVVVQFDEPTLPAALAGRLTGVTSLSPV
Mvan_2108|M.vanbaalenii_PYR-1 LTTSLAEGVAMHRAQLLRRLESPVVVQFDEPSLPAALAGRLTGVTGVSPV
MSMEG_2359|M.smegmatis_MC2_155 LGASLAEGVAAHRAEVARRLETTVVVQFDEPSLPAALQGRLSGVTSLTPV
TH_1187|M.thermoresistible__bu LAVSLAEGVAGHCAELARRLGCPVVVQFDEPSLPAASAGRLSGVTDLDSV
MAB_3352c|M.abscessus_ATCC_199 VAASLAEGLAQHRATLSRRLGVPVMVQIDEPSLPAALDGRLSGTSMFSPV
: **.**:* * * : *** *:**:** :**** * *.*.: . .*
MLBr_01706|M.leprae_Br4923 APLDETLAATLFDSCVATVGADVLLHSCAAELPWNFLQRSAIRAVSVDVN
MAV_3869|M.avium_104 PPLDEALARGLLDGCVHTVGGEVLLHCCGPDMPWNLLQDSAFSAVSVDAS
Mb3040c|M.bovis_AF2122/97 APLDETVAEALLDTCIAAVDADVALHSCSPDLPWDLLQRSRISAVSVDAS
Rv3015c|M.tuberculosis_H37Rv APLDETVAEALLDTCIAAVDADVALHSCSPDLPWDLLQRSRISAVSVDAS
MMAR_1692|M.marinum_M AALDEAVALGLLDTCVEAAGADVALHSCAAALPWDLLQRSGISAVSVDVA
MUL_1930|M.ulcerans_Agy99 AALDEAVALGLLDTCVEAAGADVALHSCAAALPWDLLQRSGISAVSVDVA
Mflv_4252|M.gilvum_PYR-GCK HPVDDAVATALLDDCVQTVGGDVVVHCCASGVPWKLLQRSAVHAVSVDVD
Mvan_2108|M.vanbaalenii_PYR-1 HPVDESVVPALLDECVEAVGGEVALHSCAADIPWKLLQRSAISAVSVDMH
MSMEG_2359|M.smegmatis_MC2_155 HPVDEPLAMSLLDACVAQADTEVAVHSCAADLPWKALLRSNIHAISVDVS
TH_1187|M.thermoresistible__bu PPVEEAVAVDLLDICSRTVGVPVAVHICAPGIPWKILQRSAIDAISIDPA
MAB_3352c|M.abscessus_ATCC_199 HPVDVPTATALLDQCAEAVGGELLVHCCASDPPWELLREANIRAIAIDVA
.:: . . *:* * .. : :* *.. **. * : . *:::*
MLBr_01706|M.leprae_Br4923 VLRTGD-LDGIAAFVESGRTVVLGVVPATAPQRLPSVEQVAAAVVGVTDR
MAV_3869|M.avium_104 ALGAAD-LDGMAAFVESGRAVALGVVAVGVPPRRPSAEQVAGAVVAVTDR
Mb3040c|M.bovis_AF2122/97 TLQAAD-LDAVAAFVESGRTVVLGLVPVTAPERAPSMEEVAAAAVAVTDR
Rv3015c|M.tuberculosis_H37Rv TLQAAD-LDAVAAFVESGRTVVLGLVPVTAPERAPSMEEVAAAAVAVTDR
MMAR_1692|M.marinum_M TLRAAD-LDSIAAFIDSGRTVVLGVVPASVPDRRPSAEEVAADVVAVTDR
MUL_1930|M.ulcerans_Agy99 TLRAAD-LDSIAAFIDSGRTVVLGVVPASVPDRRPSAEEVAADVVAVTDR
Mflv_4252|M.gilvum_PYR-GCK TLSADD-LDGVGEFVDSGRTVVLGAVPTAAPAARPSAEQIAATVAAVTDR
Mvan_2108|M.vanbaalenii_PYR-1 TLTPAD-LDGIGEFVDSNRTVLLGAVPTADPGTRPSAEQIAAAVADVTDR
MSMEG_2359|M.smegmatis_MC2_155 TLTAAD-LDGVGEFVDAGRTVLLGVVPTSAPNRRPAVEEIAKSVVAVTDR
TH_1187|M.thermoresistible__bu TLAEAD-LDGLGEFLDAGRTVQLGVVPVVMPERPPAVEEIARAAAGLTDR
MAB_3352c|M.abscessus_ATCC_199 HLADAHALDGLASVLDTGKAVVLGVIPAQAPSTPISSDVLATRTAALADK
* . **.:. .:::.::* ** :.. * : : :* .. ::*:
MLBr_01706|M.leprae_Br4923 LGFGRSALRDRIGVSPACGLAGATPHWACTAIELARKTAEAFAQDPDAI-
MAV_3869|M.avium_104 LGFTRSALRDRIGVTPACGLAGATPEWARTAIGLARRAAELFAEDPDAIE
Mb3040c|M.bovis_AF2122/97 LGVPRSALRDRLGVSPACGLANATGQWARTAVGLARDVAEAFARDPEAI-
Rv3015c|M.tuberculosis_H37Rv LGVPRSALRDRLGVSPACGLANATGQWARTAVGLARDVAEAFARDPEAI-
MMAR_1692|M.marinum_M LGFGRTVLRDRVGVAPGCGLAGATPQWARTAIGLARKAAEGLARDPDAI-
MUL_1930|M.ulcerans_Agy99 LGFGRTVLRDHVGVAPGCGLAGATPQWARTAIGLAREAAEGLARDPDAI-
Mflv_4252|M.gilvum_PYR-GCK LGFARAVLRDRVGITPACGLAGATTAWARSAIELAQRAADGLADDPEAF-
Mvan_2108|M.vanbaalenii_PYR-1 LGFPRSVLRDRVGVTPACGLAAATQAWARAAVELAQRAADGLADDPDAV-
MSMEG_2359|M.smegmatis_MC2_155 LGFAREVLRERIGVTPACGLAGATPEWARTAIELAQKVADGFAQDPEGI-
TH_1187|M.thermoresistible__bu LGFARSVLRDRIGVTPGCGLAGASGQWARTAVELARRTAEALSDDPENI-
MAB_3352c|M.abscessus_ATCC_199 LGFGRAMLRDRVGVSAGCGLAGTDLQWARKATELTRKVSDLIDADPDAL-
**. * **:::*::..**** : ** * *:: .:: : **: .