For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPTRTEIITQFVPNSPLCQHLGITLTEIGDDRAVLHMPFKSELATMGQTVHGGAIATLADTAAMAAAWAD DVVPEKFGGATASLAVNYVCAANAADLTAVGQVVRRGKRLSNIEVTVCDSAGQVVAKALATYNFA
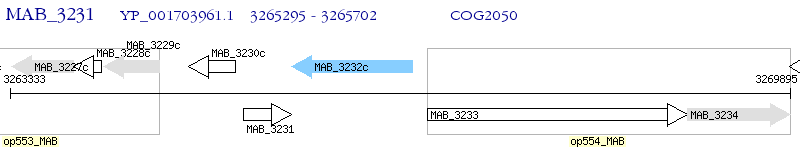
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3231 | - | - | 100% (135) | hypothetical protein MAB_3231 |
| M. abscessus ATCC 19977 | MAB_4353 | - | 3e-27 | 46.88% (128) | hypothetical protein MAB_4353 |
| M. abscessus ATCC 19977 | MAB_3925 | - | 8e-08 | 31.36% (118) | hypothetical protein MAB_3925 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1559c | - | 5e-19 | 34.11% (129) | hypothetical protein Mb1559c |
| M. gilvum PYR-GCK | Mflv_4071 | - | 1e-07 | 35.85% (106) | thioesterase superfamily protein |
| M. tuberculosis H37Rv | Rv1532c | - | 5e-19 | 34.11% (129) | hypothetical protein Rv1532c |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_4134 | - | 4e-28 | 45.67% (127) | hypothetical protein MMAR_4134 |
| M. avium 104 | MAV_1435 | - | 2e-27 | 45.24% (126) | hypothetical protein MAV_1435 |
| M. smegmatis MC2 155 | MSMEG_4984 | - | 1e-25 | 44.62% (130) | hypothetical protein MSMEG_4984 |
| M. thermoresistible (build 8) | TH_1769 | - | 2e-06 | 31.48% (108) | conserved hypothetical protein, putative |
| M. ulcerans Agy99 | MUL_3997 | - | 2e-27 | 44.88% (127) | hypothetical protein MUL_3997 |
| M. vanbaalenii PYR-1 | Mvan_2271 | - | 4e-07 | 31.78% (107) | hypothetical protein Mvan_2271 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1559c|M.bovis_AF2122/97 MSDPLTAQEQHKRRQAVRELMPRTPFIGGLGIVFERYEPDDVVIRLPFRT
Rv1532c|M.tuberculosis_H37Rv MSDPLTAQEQHKRRQAVRELMPRTPFIGGLGIVFERYEPDDVVIRLPFRT
MMAR_4134|M.marinum_M MSNASGSADNASGRDVIAQFLPQSPFVTKLGIVADHLGADEVRLRLPWDP
MUL_3997|M.ulcerans_Agy99 MSNASGSADNASGRDVIAQFLPQSPFVTKLGIVADHLGADEVRLRLPWDP
MAV_1435|M.avium_104 ---------------MIAQFLPQSPFVVKLGIVAERLDEDEVRLRLPWDP
MSMEG_4984|M.smegmatis_MC2_155 ----------MTGSDVMAQFLPTSPLVAKLGIVAENLADGQVRLRMPWDP
MAB_3231|M.abscessus_ATCC_1997 ---------MPTRTEIITQFVPNSPLCQHLGITLTEIGDDRAVLHMPFKS
Mflv_4071|M.gilvum_PYR-GCK ------------------MQVTPGHLFAQLDFRDVEDTDERMVIELDNRP
Mvan_2271|M.vanbaalenii_PYR-1 ------------------MQVTPGHLFAQMDFRDVEDSDERMIIELDNRP
TH_1769|M.thermoresistible__bu ------VTDNTEVAAPRPMTTTRDHLFGKLPFFDVVDSDERVVIDLHNRP
. : : : : : .
Mb1559c|M.bovis_AF2122/97 DLTNDGTYFHGGVIASVMDTAGAAAAWSNHDFDRGTRAATVAMSIQYTGA
Rv1532c|M.tuberculosis_H37Rv DLTNDGTYFHGGVIASVMDTAGAAAAWSNHDFDRGTRAATVAMSIQYTGA
MMAR_4134|M.marinum_M SNVTIGDMVHGGAIATLADLTVMAAAWCSGQAPPELRGVTVSLALDFMAP
MUL_3997|M.ulcerans_Agy99 SNVTIGDMVHGGAIATLADLTVMAAAWCSGQAPPELRGVTVSLALDFMAP
MAV_1435|M.avium_104 SNVTIGDMVHGGAIATLADLTVMAAAWCGAQAPPQLRGVTVSMALDFMAP
MSMEG_4984|M.smegmatis_MC2_155 SNATIADMVHGGAIATLADVTVMATAWAGAEVPGELRGVTVSMSLQYLAP
MAB_3231|M.abscessus_ATCC_1997 ELATMGQTVHGGAIATLADTAAMAAAWADDVVPEKFGGATASLAVNYVCA
Mflv_4071|M.gilvum_PYR-GCK DLVNRRGALQGGLVATLIDIA--AGRLADRHVGPGQDVTTADLTIHYLAP
Mvan_2271|M.vanbaalenii_PYR-1 DLVNRRGALQGGLVATLIDIA--AGRLADRHVGPGQDVTTADMTIHYLSP
TH_1769|M.thermoresistible__bu DLTNIRGALQGGLVATLIDIA--AGRLAVKYAGDLAAAATADMTVHYLAP
. .. .:** :*:: * : * . .*. :::.: .
Mb1559c|M.bovis_AF2122/97 AKRCDLLCHARTARRRKELTFTEITATDP-DGNIVAHAVQTYRIV---
Rv1532c|M.tuberculosis_H37Rv AKRCDLLCHARTARRRKELTFTEITATDP-DGNIVAHAVQTYRIV---
MMAR_4134|M.marinum_M ARATDVIGVGRVLRRGRSLVNCEAEIVDP-EGTLLAKALATYKVG---
MUL_3997|M.ulcerans_Agy99 ARATDVIGVGRVLRRGRSLVNCGAEIVDP-EGTLLAKALATYQVG---
MAV_1435|M.avium_104 ARASDVIGVGRVLRRGRSLVNCEAEIVDP-QGTLVAKALATYKVG---
MSMEG_4984|M.smegmatis_MC2_155 ARATDLIGVGHVLRRGHSLVHCDVDVHTP-DDTPVAKAVATYKLG---
MAB_3231|M.abscessus_ATCC_1997 ANAADLTAVGQVVRRGKRLSNIEVTVCDS-AGQVVAKALATYNFA---
Mflv_4071|M.gilvum_PYR-GCK VLEGPARAVATVVRAGRRVSVIAVDVIDVARDRLAARATVSFAVLDPR
Mvan_2271|M.vanbaalenii_PYR-1 VLDGPARAEATVVRAGRKLSVIAVDVTDVSRGRLAARATVTFAVLDPR
TH_1769|M.thermoresistible__bu IMRGPARATATLVRAGRRSIVVAVDVVDVGADRHAARATLSFAVLSVS
. * : . *:* :: .