For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVKIKLTRLGKIRNPQYRIQVADARTRREGRAIEVIGRYHPKEEPSLIEIDSERAQYWLSVGAQPTDPVL ALLKITGDWQKFKGLPGAEGTLKVKEPKPSKLDLFNAALAEADGAPTGEAIQQKKKKAPKKAEAAEAEAP AEEPAAESADAASE*
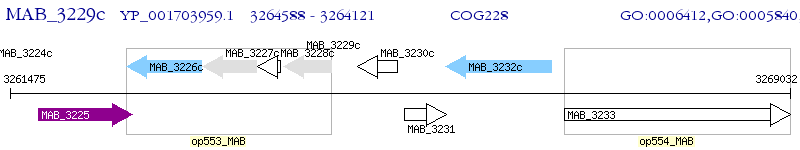
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3229c | rpsP | - | 100% (155) | 30S ribosomal protein S16 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2933c | rpsP | 2e-62 | 80.56% (144) | 30S ribosomal protein S16 |
| M. gilvum PYR-GCK | Mflv_4175 | rpsP | 3e-65 | 77.42% (155) | ribosomal protein S16 |
| M. tuberculosis H37Rv | Rv2909c | rpsP | 2e-62 | 80.56% (144) | 30S ribosomal protein S16 |
| M. leprae Br4923 | MLBr_01618 | rpsP | 1e-61 | 76.13% (155) | 30S ribosomal protein S16 |
| M. marinum M | MMAR_1799 | rpsP | 1e-64 | 78.71% (155) | 30S ribosomal protein S16 RpsP |
| M. avium 104 | MAV_3764 | rpsP | 6e-64 | 77.36% (159) | 30S ribosomal protein S16 |
| M. smegmatis MC2 155 | MSMEG_2435 | rpsP | 2e-71 | 86.45% (155) | 30S ribosomal protein S16 |
| M. thermoresistible (build 8) | TH_2390 | rpsP | 1e-65 | 76.69% (163) | PROBABLE 30S RIBOSOMAL PROTEIN S16 RPSP |
| M. ulcerans Agy99 | MUL_2047 | rpsP | 8e-65 | 78.71% (155) | 30S ribosomal protein S16 |
| M. vanbaalenii PYR-1 | Mvan_2187 | rpsP | 4e-67 | 82.47% (154) | ribosomal protein S16 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2933c|M.bovis_AF2122/97 MAVKIKLTRLGKIRNPQYRVAVADARTRRDGRAIEVIGRYHPKEEPSLIE
Rv2909c|M.tuberculosis_H37Rv MAVKIKLTRLGKIRNPQYRVAVADARTRRDGRAIEVIGRYHPKEEPSLIE
MAV_3764|M.avium_104 MAVKIKLTRLGKIRNPQYRIAVADARTRRDGRSIEIIGRYHPKEDPSLIE
MMAR_1799|M.marinum_M MAVKIKLTRLGKIRNPQYRIAVADARTRRDGRSIEVIGRYHPKEEPSLIE
MUL_2047|M.ulcerans_Agy99 MAVKIKLTRLGKIRNPQYRIAVADARTRRDGRSIEVIGRYHPKEEPSLIE
MLBr_01618|M.leprae_Br4923 MAVKIKLTRLGKIRNPQYRVIVADARTRRDGRSIEVIGRYHPKEEPSLID
MAB_3229c|M.abscessus_ATCC_199 MAVKIKLTRLGKIRNPQYRIQVADARTRREGRAIEVIGRYHPKEEPSLIE
MSMEG_2435|M.smegmatis_MC2_155 MAVKIKLTRLGKIRNPQYRIIVADARTRRDGRAIEVIGRYHPKEEPSLIQ
Mflv_4175|M.gilvum_PYR-GCK MAVKIKLARFGKIRNPQYRISVADARNRRDGRAIEVIGRYHPKEDPSIIE
Mvan_2187|M.vanbaalenii_PYR-1 MAVKIKLARFGKIRNPQYRIAVADARNRRDGRAIEVIGRYHPKEEPSVIE
TH_2390|M.thermoresistible__bu MAVKIKLTRLGKIRNPQYRIAVADARTRRDGRAIEIIGRYHPKEEPSLIE
*******:*:*********: *****.**:**:**:********:**:*:
Mb2933c|M.bovis_AF2122/97 INSERAQYWLSVGAQPTEPVLKLLKITGDWQKFKGLPGAQGRLKVAAPKP
Rv2909c|M.tuberculosis_H37Rv INSERAQYWLSVGAQPTEPVLKLLKITGDWQKFKGLPGAQGRLKVAAPKP
MAV_3764|M.avium_104 INSERAQYWLSVGAQPTEPVLKLLKITGDWQKFKGLPGAEGRLKVAPPKP
MMAR_1799|M.marinum_M INSERAQYWLSVGAQPTEPVLKLLKITGDWQKFKGLPGAEGRLKVKPPKP
MUL_2047|M.ulcerans_Agy99 INSERAQYWLSVGAQPTEPVLKLLKITGDWQKFKGLPGAEGRLKVKPPKP
MLBr_01618|M.leprae_Br4923 INSERTQYWLSVGAKPTEPVLKLLKITGDWQKFKGLPGAEGRLKVAPPKP
MAB_3229c|M.abscessus_ATCC_199 IDSERAQYWLSVGAQPTDPVLALLKITGDWQKFKGLPGAEGTLKVKEPKP
MSMEG_2435|M.smegmatis_MC2_155 IDSERAQYWLGVGAQPTEPVLALLKITGDWQKFKGLPGAEGTLKVKEPKP
Mflv_4175|M.gilvum_PYR-GCK IDSERAQYWLGVGAQPTEPVLQLLKITGDWQKFKGLPGAEGTLKHKEPKP
Mvan_2187|M.vanbaalenii_PYR-1 IDSERAQYWLGVGAQPTEPVLQLLKITGDWQKFKGLPGAEGTLKVKEPKP
TH_2390|M.thermoresistible__bu VNSERVQYWLSVGAQPTEPVLALLKLTGDWQKFKGLPGAEGSLKVKEPKP
::***.****.***:**:*** ***:*************:* ** ***
Mb2933c|M.bovis_AF2122/97 SKLEVFNAALAAADGGPTTEATKPKKKSP---AKKA----AKAAEP---A
Rv2909c|M.tuberculosis_H37Rv SKLEVFNAALAAADGGPTTEATKPKKKSP---AKKA----AKAAEP---A
MAV_3764|M.avium_104 SKLELFNAALAEAEGGPTTEAAKPKKKAATSGAKKA----AKAAEPEAAA
MMAR_1799|M.marinum_M SKLELFNAALAAAEGGPTTEATKPKKKSP---AKK-----AKGGEG---D
MUL_2047|M.ulcerans_Agy99 SKLELFNAALAAAEGGPTTEATKPKKKSP---AKK-----AKGGEG---D
MLBr_01618|M.leprae_Br4923 SKLELFNAALAVADGGPTTEATRPKKKVS---AKKA----AKAVES---D
MAB_3229c|M.abscessus_ATCC_199 SKLDLFNAALAEADGAPTGEAIQQKKKKAP--KKAE--------------
MSMEG_2435|M.smegmatis_MC2_155 SKLDLFNAALAEAESGTTAAATTPKKKKAP--KKDE--------------
Mflv_4175|M.gilvum_PYR-GCK SKLDLFNAALAEAEGGPSTEATTPKKKKAP--AKKDEQPTEKAAEPAAEK
Mvan_2187|M.vanbaalenii_PYR-1 SKLDLFNAALAEAEGGPSTEATTPKKKKAP--AKKD----------APAE
TH_2390|M.thermoresistible__bu SKLELFNQALAEAEGAPSTEAAQPKKKKAS--AKKADDKAGDKAEAKAEK
***::** *** *:...: * *** . *
Mb2933c|M.bovis_AF2122/97 PQPEQPD-------------------------TPALGGEQAELTAES
Rv2909c|M.tuberculosis_H37Rv PQPEQPD-------------------------TPALGGEQAELTAES
MAV_3764|M.avium_104 PEAAEPEAA-----------------------APAEGGEQAESSTES
MMAR_1799|M.marinum_M AD-AAAEKV-----------------------EASAEGEQTESAES-
MUL_2047|M.ulcerans_Agy99 AD-AAAEKV-----------------------EASAEGEQTESAES-
MLBr_01618|M.leprae_Br4923 AEGAKATKA-----------------------YALAEGD--EQSE--
MAB_3229c|M.abscessus_ATCC_199 AAEAEAPA-------------------------EEPAAESADAASE-
MSMEG_2435|M.smegmatis_MC2_155 AAEAPAEA-------------------------AEAPAEAADAASES
Mflv_4175|M.gilvum_PYR-GCK AAEPAAEAP------------------------AEAAAESEAPAAE-
Mvan_2187|M.vanbaalenii_PYR-1 AAEAPAEA--------------------------EAAAESDAPAAE-
TH_2390|M.thermoresistible__bu KADEKAEKKADEKAEKKAGEKAEKKAGEKAGEKAEAAAEGSDAAKES
. .: :