For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQEREPTLWAISDLHVGQSANKSVLEELHPASPEDWLIVAGDVAERSDDIRAALDLLRRRFTKVIWVPGN HELWTTARDPMQIFGQSRYDYLVNMCDEMGVITPEHPYPVWVEQGGPATIVPMFVLYDYTFLPEGANSKA EGLKIARDRNVVATDEFLLSSEPFATRDAWCRDRLRYTRKRLEDLDWMTPTVLVNHFPLVREPCDALFYP EFSLWCGTTETADWHTRYNAACSVYGHLHIPRTTWYDDVRFEEVSVGYPREWAQRKPYSWLRQILPAPHY PPGYLNEFGGHFEITEQMRVAAKRFRERLAKERASD*
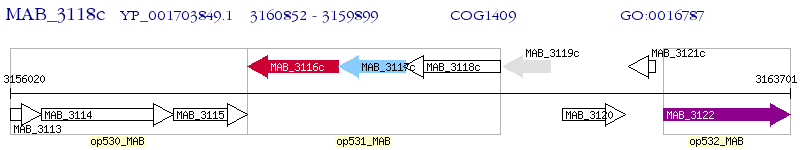
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3118c | - | - | 100% (317) | hypothetical protein MAB_3118c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2818c | - | 1e-158 | 79.68% (310) | hypothetical protein Mb2818c |
| M. gilvum PYR-GCK | Mflv_4030 | - | 1e-159 | 78.64% (309) | metallophosphoesterase |
| M. tuberculosis H37Rv | Rv2795c | - | 1e-158 | 79.68% (310) | hypothetical protein Rv2795c |
| M. leprae Br4923 | MLBr_01548 | - | 1e-156 | 78.71% (310) | hypothetical protein MLBr_01548 |
| M. marinum M | MMAR_1915 | - | 1e-156 | 78.64% (309) | hypothetical protein MMAR_1915 |
| M. avium 104 | MAV_3684 | - | 1e-161 | 81.94% (310) | metallophosphoesterase |
| M. smegmatis MC2 155 | MSMEG_2647 | - | 1e-161 | 79.23% (313) | metallophosphoesterase |
| M. thermoresistible (build 8) | TH_1426 | - | 1e-155 | 77.64% (313) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2140 | - | 1e-155 | 78.76% (306) | hypothetical protein MUL_2140 |
| M. vanbaalenii PYR-1 | Mvan_2327 | - | 1e-159 | 78.03% (314) | metallophosphoesterase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2818c|M.bovis_AF2122/97 MTWKGSGQETVGAEPTLWAISDLHTGHLGNKPVAESLYPSSPDDWLIVAG
Rv2795c|M.tuberculosis_H37Rv MTWKGSGQETVGAEPTLWAISDLHTGHLGNKPVAESLYPSSPDDWLIVAG
MMAR_1915|M.marinum_M MTGHES---TGGAQPTLWAVSDLHTGHMGNKPVTESLYPSSPEDWLIVAG
MUL_2140|M.ulcerans_Agy99 ----------------MWAVSDLHTGHMGNKPVTESLYPSSPEDWLIVAG
MLBr_01548|M.leprae_Br4923 MAKQEP---MCGVEPTLWAISDLHTGHVGNKPVAESLYPLSPDDWLIVAG
MAV_3684|M.avium_104 MAGQES---SNAGQPTLWAVSDLHTGHLGNKPVTESLHPSSPEDWLIVAG
Mflv_4030|M.gilvum_PYR-GCK MSTDE-------RRPTLWAVSDLHTGHTGNKPVTESLHPASPDDWLIVAG
Mvan_2327|M.vanbaalenii_PYR-1 MTTDE-------RRPTLWAVSDLHTGHTGNKPVTESLHPASPDDWLIVAG
MSMEG_2647|M.smegmatis_MC2_155 MVTDHDS---RDRQPVLWAISDLHTGHTGNKPVTESLYPATPDDWLIVAG
TH_1426|M.thermoresistible__bu MGQDQD----RSRRPVLWAISDLHTGHTGNRPVTESLYPSTPDDWLIVAG
MAB_3118c|M.abscessus_ATCC_199 MTQER--------EPTLWAISDLHVGQSANKSVLEELHPASPEDWLIVAG
:**:****.*: .*:.* *.*:* :*:*******
Mb2818c|M.bovis_AF2122/97 DVAERTDEIRWSLDLLRRRFAKVIWVPGNHELWTTNRDPMQIFGRARYDY
Rv2795c|M.tuberculosis_H37Rv DVAERTDEIRWSLDLLRRRFAKVIWVPGNHELWTTNRDPMQIFGRARYDY
MMAR_1915|M.marinum_M DVAERTDEIRWSLDLLRRRFAKVIWVPGNHELWTTTRDPMQIFGRARYDY
MUL_2140|M.ulcerans_Agy99 DVAERTDEIRWSLDLLRRRFAKVIWVPGNHELWTTTRDPMQIFGRARYDY
MLBr_01548|M.leprae_Br4923 DVAECTDEIRWTLELLRHRFAKVIWVPGNHELWTTNRDPMQIFGRARYDY
MAV_3684|M.avium_104 DVAERTDDIRWALDLLRRRFAKVIWVPGNHELWTTTRDPVQVFGKARYDY
Mflv_4030|M.gilvum_PYR-GCK DVAERTDEIRWALDLLRKRFAKVIWIPGNHELWTTQRDPMQIYGKSRYDY
Mvan_2327|M.vanbaalenii_PYR-1 DVGERTDEIRWALDLLRKRFAKVIWVPGNHELWTTQRDPMQIFGRARYDY
MSMEG_2647|M.smegmatis_MC2_155 DVGERTDEIRWALDLLRKRFAKVIWVPGNHELWTTNKDPMQIFGRARYDY
TH_1426|M.thermoresistible__bu DVAERTDEIRWALDVLRKRFAKVIWVPGNHELWTTSRDPVQVFGRSRYDY
MAB_3118c|M.abscessus_ATCC_199 DVAERSDDIRAALDLLRRRFTKVIWVPGNHELWTTARDPMQIFGQSRYDY
**.* :*:** :*::**:**:****:********* :**:*::*::****
Mb2818c|M.bovis_AF2122/97 LVNMCDEMGVVTPEHPFPVWTERGGPATIVPMFLLYDYSFLPEGANSKAE
Rv2795c|M.tuberculosis_H37Rv LVNMCDEMGVVTPEHPFPVWTERGGPATIVPMFLLYDYSFLPEGANSKAE
MMAR_1915|M.marinum_M LVNMCDEMGVVTPEHPFPVWTERGGPATIVPMFLLYDYSFLPSGATTKAE
MUL_2140|M.ulcerans_Agy99 LVNMCDEMGVVTPEHPFPVWTERGGPATIVPMFLLYDYSFLPSGATTKAE
MLBr_01548|M.leprae_Br4923 LINMCDQMGVVTSEHPFPLWTERGGPATIVPMFLLYDYTFLPTGADSKAK
MAV_3684|M.avium_104 LVNMCDEMGVITPEHPFPVWTERGGPATIVPMFLLYDYSFLPDGAASKAE
Mflv_4030|M.gilvum_PYR-GCK LVTMCDEMGIVTPEHPYPVWTEEGGPATIVPMFLLYDYSFLPQGAGTKAE
Mvan_2327|M.vanbaalenii_PYR-1 LVNMCDEMGIVTPEHPYPVWTEEGGPATIVPMFLLYDYSFLPQGAGTKAE
MSMEG_2647|M.smegmatis_MC2_155 LVDMCDQMGVVTPEHPFPVWTEQGGPATIVPMFLLYDYSFLPEGASSKAE
TH_1426|M.thermoresistible__bu LVEMCDKMGIITPEHSYPVWTEEGGPATIVPMFLLYDYSFLPAGAATKEE
MAB_3118c|M.abscessus_ATCC_199 LVNMCDEMGVITPEHPYPVWVEQGGPATIVPMFVLYDYTFLPEGANSKAE
*: ***:**::*.**.:*:*.*.**********:****:*** ** :* :
Mb2818c|M.bovis_AF2122/97 GVAIAKERNVVATDEFLLSPEPYPTRDAWCHERVAATRARLEQLDWMQPT
Rv2795c|M.tuberculosis_H37Rv GVAIAKERNVVATDEFLLSPEPYPTRDAWCHERVAATRARLEQLDWMQPT
MMAR_1915|M.marinum_M GMAIAKQRNVVATDEFLLSPEPYATRDAWCRERVAATRARLEQLDWMQPT
MUL_2140|M.ulcerans_Agy99 GMAIAKQRNVVATDEFLLSPEPYATRDAWCRERFAATRARLEQLDWMQPT
MLBr_01548|M.leprae_Br4923 GLAIARERNVVATDEYLLSSEPYATREAWCRDRLDVTRSRLEQLDWMTPT
MAV_3684|M.avium_104 GLMIARDRNVVATDEFLLSPEPYPTREAWCRERLEITRARLEALDWMTPT
Mflv_4030|M.gilvum_PYR-GCK GLAIAREKNIVGTDEYLLSSEPYATRDAWCRDRVNYTRGRLEDLDWMMPT
Mvan_2327|M.vanbaalenii_PYR-1 GLAIAKERNIVGTDEYLLSAEPYSTRDAWCHDRVNYTRKRLEDLDWMMPT
MSMEG_2647|M.smegmatis_MC2_155 GLAIAKERNIVGTDEFLLSCEPYATRDAWCRDRVAHTRKRLEDLDWMTPT
TH_1426|M.thermoresistible__bu GLAMARENNVVATDEFLLSSEPYATRDAWCRERVAETRKRLEQLDWMTPT
MAB_3118c|M.abscessus_ATCC_199 GLKIARDRNVVATDEFLLSSEPFATRDAWCRDRLRYTRKRLEDLDWMTPT
*: :*::.*:*.***:*** **:.**:***::*. ** *** **** **
Mb2818c|M.bovis_AF2122/97 VLVNHFPLLRQPCDALFYPEFSLWCGTTKTADWHTRYNAVCSVYGHLHIP
Rv2795c|M.tuberculosis_H37Rv VLVNHFPLLRQPCDALFYPEFSLWCGTTKTADWHTRYNAVCSVYGHLHIP
MMAR_1915|M.marinum_M VLVNHFPLLREPCDAMFYPEFSMWCGTTKTADWHTRYNALCSVYGHLHIP
MUL_2140|M.ulcerans_Agy99 VLVNHFPLLREPCDAMFYPEFSMWCGTTKTADWHTRYNALCSVYGHLHIP
MLBr_01548|M.leprae_Br4923 VLVNHFPLVREPCDALFYPEFSLWCGTTKTADWHIRYNAVCSVYGHLHIP
MAV_3684|M.avium_104 VLVNHFPLVREPCDALFYPEFSLWCGTTKTADWHTRYNAVCSVYGHLHIP
Mflv_4030|M.gilvum_PYR-GCK IQVNHFPMVREPCDAMFYPEFSLWCGTTATADWHTRYNAICSVYGHLHIP
Mvan_2327|M.vanbaalenii_PYR-1 VQVNHFPMVREPCDAMFYPEFSLWCGTTATADWHTRYNAVCSVYGHLHIP
MSMEG_2647|M.smegmatis_MC2_155 VLVNHFPMVREPCDAMFYPEFSLWCGTTATADWHTRYNAVCSVYGHLHIP
TH_1426|M.thermoresistible__bu VLVNHFPLVREPCDALFYPEFSLWCGTTATADWHTRYNAVCSVYGHLHIP
MAB_3118c|M.abscessus_ATCC_199 VLVNHFPLVREPCDALFYPEFSLWCGTTETADWHTRYNAACSVYGHLHIP
: *****::*:****:******:***** ***** **** **********
Mb2818c|M.bovis_AF2122/97 RTTWYDGVRFEEVSVGYPREWRRRKPYSWLRQVLPDPQYAPGYLNDFGGH
Rv2795c|M.tuberculosis_H37Rv RTTWYDGVRFEEVSVGYPREWRRRKPYSWLRQVLPDPQYAPGYLNDFGGH
MMAR_1915|M.marinum_M RTTWHDGVRFEEVSVGYPREWRRRKPYSWLRQVLPDPQYAPGYLNDFGGH
MUL_2140|M.ulcerans_Agy99 RTTWHDGVRFEEVSVGYPREWRRRKPYSWLRQVLPDPQYAPGYLNDFGGH
MLBr_01548|M.leprae_Br4923 RTTWYNEVRFEEVSVGYPREWRRRKPYSWLRQVLPDPQYAPGYLNDFGGH
MAV_3684|M.avium_104 RTTWYDDVRFEEVSVGYPREWRRRKPYGWLRQVLPDPQYAPGYLNDFGGH
Mflv_4030|M.gilvum_PYR-GCK RTTWYDGVRFEEVSVGYPREWRRRRPYRWLRQILPDPKYPAGYLNEFGGH
Mvan_2327|M.vanbaalenii_PYR-1 RTTWYDGVRFEEVSVGYPREWRRRRPYRWLRQILPDPKYAPGYLNEFGGH
MSMEG_2647|M.smegmatis_MC2_155 RTTWYDDVRFEEVSVGYPREWRRRKPYRWLRQILPDPKYAPGYLNEFGGH
TH_1426|M.thermoresistible__bu RTTWYDGVRFEEVSVGYPREWRRRKPYRWLRQVLPDPQYAPGYLNEFGGH
MAB_3118c|M.abscessus_ATCC_199 RTTWYDDVRFEEVSVGYPREWAQRKPYSWLRQILPAPHYPPGYLNEFGGH
****:: ************** :*:** ****:** *:*..****:****
Mb2818c|M.bovis_AF2122/97 FVITPEMRTQAAQFRERLRQRQSR-
Rv2795c|M.tuberculosis_H37Rv FVITPEMRTQAAQFRERLRQRQSR-
MMAR_1915|M.marinum_M FVITPEMRAQATKFRERLRQRQER-
MUL_2140|M.ulcerans_Agy99 FVITPEMRAQATKFRERLRQRQER-
MLBr_01548|M.leprae_Br4923 FVITSEMRAQAVQFRERLRQKQSR-
MAV_3684|M.avium_104 FEITPEMRRQATQFRERLRQRQSR-
Mflv_4030|M.gilvum_PYR-GCK FQITQEMRDNAAKMQQRIKDRG---
Mvan_2327|M.vanbaalenii_PYR-1 FQITQEMRDNAEKMQQRIRDRA---
MSMEG_2647|M.smegmatis_MC2_155 FEITPEMRANAQQMQERIKSRRGL-
TH_1426|M.thermoresistible__bu FVITAEMRANAQRVQERIQARRARR
MAB_3118c|M.abscessus_ATCC_199 FEITEQMRVAAKRFRERLAKERASD
* ** :** * :.::*: .