For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DKRAAAVQDARMIDRAGLAGFLRSRREAMQPDDVGLPRGQRRRTRGLRREEVAALCHMSTDYYARLEGAR GPQPSEQMIAAIAQGLHLSREERDHLFRLAGHQPPASGGESEYISPGLMRIFDRLTPDTAAEIVTELGET LRQTPLGIALAGDLTRYAGPARSIGYRWFTDPATRSIHPVEDHQFYSRMYASGLRRVLAARGPGSKAARF AELLNAESEQFRALWKEHEVGVMPKEMKRFRHPEVGLLELNCQILLDPVESTSLLVYTAVPGSESYEKLQ LLAIIGASSPPGR*
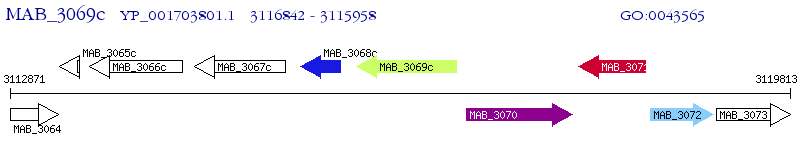
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3069c | - | - | 100% (294) | putative DNA-binding protein |
| M. abscessus ATCC 19977 | MAB_4641 | - | 5e-25 | 32.23% (273) | putative DNA-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5170 | - | 2e-26 | 31.64% (275) | XRE family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2211 | - | 1e-111 | 69.42% (278) | DNA-binding protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1062 | - | 1e-76 | 52.31% (281) | XRE family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_3069c|M.abscessus_ATCC_199 MDKRAAAVQDARMIDRAGLAGFLRSRREAMQPDDVGLPRGQRRRTRGLRR
MSMEG_2211|M.smegmatis_MC2_155 ------------MIDRTGLAEFLRHRRESLQPEDVGLPRGRRRRTNGLRR
Mvan_1062|M.vanbaalenii_PYR-1 -------------MDQRQLADFLRTRREALTPADVGLPTGRRRRTAGLRR
Mflv_5170|M.gilvum_PYR-GCK --------MDTRAQRRLALGEFLRARREAIRRADVGLPELPRARTGGLRR
: *. *** ***:: ***** * ** ****
MAB_3069c|M.abscessus_ATCC_199 EEVAALCHMSTDYYARLEGARGPQPSEQMIAAIAQGLHLSREERDHLFRL
MSMEG_2211|M.smegmatis_MC2_155 EEVAALCHMSTDYYSRLERGHGPKPSEQMIASMAQGLHLSLDERDHLFRL
Mvan_1062|M.vanbaalenii_PYR-1 EEVAALAMISTDYYSRMEQQRGPQPSVDVLASLARALQLTLDERDHLFRL
Mflv_5170|M.gilvum_PYR-GCK EEVAVTAGVSVTWYTWLEQGRDINPSKQVLDAVADALRLSPAEHEYVLAL
****. . :*. :*: :* :. :** ::: ::* .*:*: *::::: *
MAB_3069c|M.abscessus_ATCC_199 AGHQPPASGGESEYISPGLMRIFDRLTPDTAAEIVTELGETLRQTPLGIA
MSMEG_2211|M.smegmatis_MC2_155 AGCSPPARGTDSEHISPGMLRIFDRLD-DTPAEIVTELGETLRQTPLSVA
Mvan_1062|M.vanbaalenii_PYR-1 AGHNAPARTPAGEQVSTAMLRILDRLS-DTPAQVMSALGETLVQTPAARA
Mflv_5170|M.gilvum_PYR-GCK GGYAVDAAVSRG-PLPPHIQHFLDALEGYPAFAITPDWGIAAWNT-TYAA
.* * . :.. : :::* * .. : . * : :* *
MAB_3069c|M.abscessus_ATCC_199 LAGDLTRYAGPARSIGYRWFTDPATRSIHPVEDHQFYSRMYASGLRRVLA
MSMEG_2211|M.smegmatis_MC2_155 LTGDLTVHTGPARSIGYRWFTDEATRRLYAPEEHDFLTRMFVSNLRALVA
Mvan_1062|M.vanbaalenii_PYR-1 LLGDETTYRGLARSVVYRWFTDPTSRSVYPAEDHAAIGRNYVAAARMGYT
Mflv_5170|M.gilvum_PYR-GCK LYPNVERVPETDRNLLWLVFTDPYIHELLPNWDTD--SRRFLAEFRADAG
* : *.: : *** : : . : * : : *
MAB_3069c|M.abscessus_ATCC_199 ARGPGSKAARFAELLNAESEQFRALWKEHEVGVMPKEMKRFRHPEVGLLE
MSMEG_2211|M.smegmatis_MC2_155 LRGAGSRAAHYADLLLSESDEFRRVWDDHEIGLRPREVKHFVHPELGTLE
Mvan_1062|M.vanbaalenii_PYR-1 RDGAGSRAAEIVDDLLSRSDEFAELWKQHLVGTAHELDKRLVHPALGVLE
Mflv_5170|M.gilvum_PYR-GCK PRVGEPSYTALVSRLSSESPHFAAAWQATSIERFTSRERLFRHPAAGELR
. : .. * :.* .* *. : : : ** * *.
MAB_3069c|M.abscessus_ATCC_199 LNCQILLDPVE-STSLLVYTAVPGSESYEKLQLLAIIGASSPPGR-----
MSMEG_2211|M.smegmatis_MC2_155 LHCQTLLDPEQ-SHLLLVYTAVPGSESREKLQLLSVIGAQALR-------
Mvan_1062|M.vanbaalenii_PYR-1 LQCQILHDPAQ-LHTLLVFTAEPGTPSHAKLAALGADPQSYRRGRSINGS
Mflv_5170|M.gilvum_PYR-GCK LEHHQVAPVDVPDIQIVAYLPVPDSVASERLRMLTGQADTAR--------
*. : : ::.: . *.: : :* *
MAB_3069c|M.abscessus_ATCC_199 ----
MSMEG_2211|M.smegmatis_MC2_155 ----
Mvan_1062|M.vanbaalenii_PYR-1 TVPE
Mflv_5170|M.gilvum_PYR-GCK ----