For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPGTGSDDDFINRAFGPALTQVGATLIAVRPEPDDLVNGYRRALNQAAQLGPIVVGGVSIGTAVALTWAL QNPEATVAVLAAMPAWTGAPENSPASVSARVTAESLRSDGLAAVTAAMQASSPAWLAAELTRSWAVQWPG LPHAMEEVSAFVNPSAAEIRALSVPMAVAAATDDPIHPLEVGRDWAAWAPRAALRTFTLDQLGADPACLG RACIAALADI
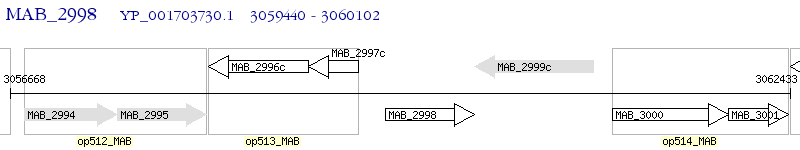
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2998 | - | - | 100% (220) | hypothetical protein MAB_2998 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2714 | - | 3e-77 | 61.82% (220) | hypothetical protein Mb2714 |
| M. gilvum PYR-GCK | Mflv_3931 | - | 5e-69 | 55.86% (222) | hypothetical protein Mflv_3931 |
| M. tuberculosis H37Rv | Rv2695 | - | 3e-77 | 61.82% (220) | hypothetical protein Rv2695 |
| M. leprae Br4923 | MLBr_01030 | - | 2e-74 | 60.55% (218) | hypothetical protein MLBr_01030 |
| M. marinum M | MMAR_2019 | - | 8e-75 | 61.36% (220) | hypothetical protein MMAR_2019 |
| M. avium 104 | MAV_3587 | - | 2e-81 | 65.91% (220) | hypothetical protein MAV_3587 |
| M. smegmatis MC2 155 | MSMEG_2767 | - | 1e-73 | 60.00% (220) | hypothetical protein MSMEG_2767 |
| M. thermoresistible (build 8) | TH_1194 | - | 2e-42 | 57.89% (133) | CONSERVED HYPOTHETICAL ALANINE RICH PROTEIN |
| M. ulcerans Agy99 | MUL_3333 | - | 1e-74 | 60.91% (220) | hypothetical protein MUL_3333 |
| M. vanbaalenii PYR-1 | Mvan_2469 | - | 5e-71 | 57.53% (219) | hypothetical protein Mvan_2469 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3931|M.gilvum_PYR-GCK -------MTIDLRGVTAVLLPGTGSDDDFAYRAFATALHEAGAVVVTPRP
Mvan_2469|M.vanbaalenii_PYR-1 -------MAVDLRGVTTVLLPGTGSDDDFVYRAFAPALHEAGAIVVTPPP
Mb2714|M.bovis_AF2122/97 -------MAVDLDGVTTVLLPGTGSDNDYVRRAFSAPLRRAGAVLVTPVP
Rv2695|M.tuberculosis_H37Rv -------MAVDLDGVTTVLLPGTGSDNDYVRRAFSAPLRRAGAVLVTPVP
MLBr_01030|M.leprae_Br4923 -------MAVDLRNVTTVLLPGTGSDDDYVYRAFSGPLHRVGAAVLTPPP
MAV_3587|M.avium_104 -------MVVDLRGVSAVLLPGTGSDDDYIHRAFSGPLTGVGAVLVAPPP
MMAR_2019|M.marinum_M MTGHYRGVPVDLRGVPVALLPGTGSDDDYVYRAFSGPLQSVEAILVTPPP
MUL_3333|M.ulcerans_Agy99 -------MPVDLRGVPVALLPGTGSDDDYVYRAFSGPLQSVEAILVTPPP
MAB_2998|M.abscessus_ATCC_1997 -------------------MPGTGSDDDFINRAFGPALTQVGATLIAVRP
MSMEG_2767|M.smegmatis_MC2_155 ------MMSVILRGVTTVLLPGTGSDDDYVYRAFSTALHEVGAPLVTPPP
TH_1194|M.thermoresistible__bu ---------------------------------------------VWPAG
:
Mflv_3931|M.gilvum_PYR-GCK EPQRLIDGYREQFDSAWHACAPQQIVVGGVSIGAAVAVEWAVAHPGCALA
Mvan_2469|M.vanbaalenii_PYR-1 QPHRLIDGYRDELR---DAARSQPIAVGGVSIGAAVAVEWALAHPTQVVA
Mb2714|M.bovis_AF2122/97 HPGRLIDGYRAALD---DAARDGPVVVGGVSLGAAVAAAWALEHPDRAVA
Rv2695|M.tuberculosis_H37Rv HPGRLIDGYRAALD---DAARDGPVVVGGVSLGAAVAAAWALEHPDRAVA
MLBr_01030|M.leprae_Br4923 QPNRLIDGYLSALD---DAARAGPIGVGGVSIGAAVAAAWALAHPERAVA
MAV_3587|M.avium_104 HPDRLVAGYLEALD---DAARRGPIAVGGVSIGAAVAAAWALAHPDRAVA
MMAR_2019|M.marinum_M QPEHLIEGYLAALD---EAAQAGPIAVGGVSIGAAVAVAWALAHPDRCVA
MUL_3333|M.ulcerans_Agy99 QPEHLIEGYLAALD---EAAQAGPIAVGGVSIGAAVAVAWALAHPDRCVA
MAB_2998|M.abscessus_ATCC_1997 EPDDLVNGYRRALN---QAAQLGPIVVGGVSIGTAVALTWALQNPEATVA
MSMEG_2767|M.smegmatis_MC2_155 QPGRLVAGYRDALD---NAARSGPIAVGGVSIGAVVALRWALNHPNAAVA
TH_1194|M.thermoresistible__bu TTDWVMCNARRGFS--------GETSCRRADPGPPVTR------------
. :: . : .. *. *:
Mflv_3931|M.gilvum_PYR-GCK VLAALPAWSGDPDTAPAALAARYSAEQLRRDGLGTTTAQMRAGSPAWLAD
Mvan_2469|M.vanbaalenii_PYR-1 VLAALPAWTGAPDAAPAAMAARHSAELLRRDGLVATTTQMQATSPRWLAE
Mb2714|M.bovis_AF2122/97 VLAALPAWTGEPELAPAAQAARYTAARLRCDGLAATTTRMRASSPVWLAE
Rv2695|M.tuberculosis_H37Rv VLAALPAWTGEPELAPAAQAARYTAARLRCDGLAATTTRMRASSPVWLAE
MLBr_01030|M.leprae_Br4923 VLAALPAWNGAPESAPAALAARYSASHLRRDGLAATTLQMQASSPPWLAD
MAV_3587|M.avium_104 VLAALPAWAGAPEEAPAALAARYSAARLRADGLAATTIQMRASSPAWLAD
MMAR_2019|M.marinum_M VLAALPAWTGEPGPAPAAHAARYTAAQLRRDGLATTTMQMRASSPPWLAD
MUL_3333|M.ulcerans_Agy99 VLAALPAWTGEPGPAPAAHAARYTAAQLRRDGLATTTTQMRASSPPWLAD
MAB_2998|M.abscessus_ATCC_1997 VLAAMPAWTGAPENSPASVSARVTAESLRSDGLAAVTAAMQASSPAWLAA
MSMEG_2767|M.smegmatis_MC2_155 VLAALPPWSGAPHDAPAALLARQSALSLRRDGLGATVAEMRRNSPAWLAD
TH_1194|M.thermoresistible__bu --------CGGPXXXPAAAAARYSAHRLREDGLAGATAQMRATSPAWLAE
* * **: ** :* ** *** .. *: ** ***
Mflv_3931|M.gilvum_PYR-GCK ELTRSWAGQWPALPDAMDEAGRYVAPTSAALETLTAPLGVVSATDDAVHP
Mvan_2469|M.vanbaalenii_PYR-1 ELTRSWLSQWPALPEAMDAAAGYVAPTSGEMERLGAALGVVSAVDDPVHP
Mb2714|M.bovis_AF2122/97 ELTRSWRVQWPELPDAMEEAAAYVAPSRAELARLVAPLAVAAAVDDPIHP
Rv2695|M.tuberculosis_H37Rv ELTRSWRVQWPELPDAMEEAAAYVAPSRAELARLVAPLAVAAAVDDPIHP
MLBr_01030|M.leprae_Br4923 ELARSWCGQWPLLPDAMEEAAAYIAPSCAELARLATPLGVAAAVDDPIHP
MAV_3587|M.avium_104 ELTRSWGAQWPHLPDAMEEAAGYVAPSRDELAGLTAPLAVAAALDDPIHP
MMAR_2019|M.marinum_M ELARSWRVQWPQLPDAMEQAADYVAPSSTELGRLTVPLGVAAATDDPIHP
MUL_3333|M.ulcerans_Agy99 ELARSWRVQWPQLPDAMEQAADYVAPSSTELGRLTVPLGVAAATDDPIHP
MAB_2998|M.abscessus_ATCC_1997 ELTRSWAVQWPGLPHAMEEVSAFVNPSAAEIRALSVPMAVAAATDDPIHP
MSMEG_2767|M.smegmatis_MC2_155 ELARSWLGQWPALPESMEEAAAYHAPECAELERLTVPMGVAVATDDPVHP
TH_1194|M.thermoresistible__bu ELSRSWAAQWPALPEAMEAAAGYVAPTTAELARLTVPLAAVAAEDDPVHP
**:*** *** **.:*: .. : * : * ..:... * **.:**
Mflv_3931|M.gilvum_PYR-GCK LEVGVEWVSAAPRAALRTVTLDDIGADPGILGRLSLAAVQDAQSRS----
Mvan_2469|M.vanbaalenii_PYR-1 LEVGIEWVSAAPRAALRTLTLNAIGADPAVLGAESLAALWDADNPARPAV
Mb2714|M.bovis_AF2122/97 LQVAADWVSVAPHAALRTVTLDEIGADAAALGSACLAALAEVSGA-----
Rv2695|M.tuberculosis_H37Rv LQVAADWVSVAPHAALRTVTLDEIGADAAALGSACLAALAEVSGA-----
MLBr_01030|M.leprae_Br4923 LQVGVDWVTAAPHAALQTVTLNQIGTNAAALGTACLAALALT--------
MAV_3587|M.avium_104 LQAGRDWVAAAPRAALRTVTLDQIGADPAALGAACLAALAEL--------
MMAR_2019|M.marinum_M LQVGLDWASAAAHAALRTVTLEQMGADTSALGAACLAALADI--------
MUL_3333|M.ulcerans_Agy99 LQVGLDWVSAAAHAALRTVTLEQMGADTSALGAACLAALADI--------
MAB_2998|M.abscessus_ATCC_1997 LEVGRDWAAWAPRAALRTFTLDQLGADPACLGRACIAALADI--------
MSMEG_2767|M.smegmatis_MC2_155 AEVGYAWVSAAPRAALRTVTLEQMGIDTGSLGAACVAALGDV--------
TH_1194|M.thermoresistible__bu IEVARAWVAAAPRAALRTVTLAEMGADPSVLGAGCVAALQGVADARTATA
:.. *.: *.:***:*.** :* :.. ** .:**:
Mflv_3931|M.gilvum_PYR-GCK ------
Mvan_2469|M.vanbaalenii_PYR-1 R-----
Mb2714|M.bovis_AF2122/97 ------
Rv2695|M.tuberculosis_H37Rv ------
MLBr_01030|M.leprae_Br4923 ------
MAV_3587|M.avium_104 ------
MMAR_2019|M.marinum_M ------
MUL_3333|M.ulcerans_Agy99 ------
MAB_2998|M.abscessus_ATCC_1997 ------
MSMEG_2767|M.smegmatis_MC2_155 ------
TH_1194|M.thermoresistible__bu VEGAER