For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDPQSGAALCLQHLADRHPTVSNERLIAQLVPPPTFSSVSFDSYRPDPNEPTQTAAVEACRAFAQEATTR RAGKKKLFGKREVLPGVGIYLDGGFGVGKTHLLASIYHTVPGPKAFASFGELTQVASVFGFLECIELLAD YTVVCIDEFELDDPGNTTLVSRLLSELVARGVSIAATSNTLPEQLGEGRFAAQDFLREIAALSAIFNTVR VEGPDYRHRDLPPAPEPHSDEQLRELAGAINGATFDRFDDLCKHLASMHPSRYLTLIEGVTAVFIADVEQ VHDQSVALRIVALTDRLYDAGIPVQASGAKLNTIFDDEMVAGGFRKKYLRATSRLLALSAAPLPNAA*
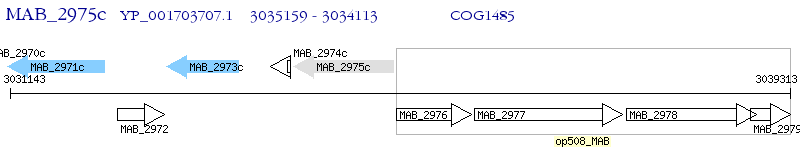
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2975c | - | - | 100% (348) | AFG1-like ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2689c | - | 1e-143 | 73.26% (344) | hypothetical protein Mb2689c |
| M. gilvum PYR-GCK | Mflv_3912 | - | 1e-146 | 74.33% (335) | AFG1 family ATPase |
| M. tuberculosis H37Rv | Rv2670c | - | 1e-143 | 73.26% (344) | hypothetical protein Rv2670c |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2046 | - | 1e-143 | 72.67% (344) | hypothetical protein MMAR_2046 |
| M. avium 104 | MAV_3561 | - | 1e-141 | 70.89% (347) | ATP/GTP-binding integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_2788 | - | 1e-144 | 76.44% (331) | ATP/GTP-binding integral membrane protein |
| M. thermoresistible (build 8) | TH_2402 | - | 1e-149 | 78.53% (326) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3304 | - | 1e-142 | 72.09% (344) | hypothetical protein MUL_3304 |
| M. vanbaalenii PYR-1 | Mvan_2491 | - | 1e-147 | 76.29% (329) | AFG1 family ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3912|M.gilvum_PYR-GCK ------------MHG------SSDVEHLVDRHPSVTPERLVAQLIPPPTF
Mvan_2491|M.vanbaalenii_PYR-1 ------------MHG------SSDVAHLVDRQPSVTPERLIAQLIPPPTF
MSMEG_2788|M.smegmatis_MC2_155 ------------MHG------SSGVASLSDRRPSVTPERLVAELVPPPTF
TH_2402|M.thermoresistible__bu ----------------------------VDRHPSVTPERLIAQLVPPPTF
Mb2689c|M.bovis_AF2122/97 MTLIAARRYSATMHGSASE-ACGSVDHLVDRHPTVSPVRLIAQLRPPPTF
Rv2670c|M.tuberculosis_H37Rv MTLIAARRYSATMHGSASE-ACGSVDHLVDRHPTVSPVRLIAQLRPPPTF
MMAR_2046|M.marinum_M ------------MHGSTSAGAWGGVAHLVDRHPSVSPERLIAQLRPPPTF
MUL_3304|M.ulcerans_Agy99 MKLIATRRYSASMHGSTSAGAWGGVAHLVDRHPSVSPERLIAQLRPPPTF
MAV_3561|M.avium_104 ------------MHGSGSASASAGVAHLVDRHPTVSPQRLIAQLRPPPTF
MAB_2975c|M.abscessus_ATCC_199 ------------MSDPQSG-AALCLQHLADRHPTVSNERLIAQLVPPPTF
**:*:*: **:*:* *****
Mflv_3912|M.gilvum_PYR-GCK ADVSFDTYRPDPSEPSQAAAVQTCRKFAEQAVERRAGKKKLFGKREVLPG
Mvan_2491|M.vanbaalenii_PYR-1 ADVSFDSYRPDPAEPSQAAAVQTCREFCEQAEQRRAGRKKLFGKREVLPG
MSMEG_2788|M.smegmatis_MC2_155 ADVSFDSYRPDPAEPTQAAAVQSCRQFCEQAAERRAGKKKLFGKREVLPG
TH_2402|M.thermoresistible__bu ADVSFDSYRPDPAEPSQAAAVRACREFCEQAVTRRAGKKRLFGKRTVLPG
Mb2689c|M.bovis_AF2122/97 AEVSFATYRPDPVEPTQAAAVVACQDFCRQAVERRAGRKKWFGKRDVLPG
Rv2670c|M.tuberculosis_H37Rv AEVSFATYRPDPVEPTQAAAVVACQDFCRQAVERRAGRKKWFGKRDVLPG
MMAR_2046|M.marinum_M AEVSFSTYRPDPAEPTQAAALVACRDFCQEAVQRRAGRKKLLGRREVSPG
MUL_3304|M.ulcerans_Agy99 AEVSFSTYRPDPAEPTQAAALVACRDFCQEAVQRRAGRKKLLGRREVLPG
MAV_3561|M.avium_104 ADVSFATYQPDPAEPTQAAAVAACREFCRQAVQRRAGRRKLLGRREILPG
MAB_2975c|M.abscessus_ATCC_199 SSVSFDSYRPDPNEPTQTAAVEACRAFAQEATTRRAGKKKLFGKREVLPG
:.*** :*:*** **:*:**: :*: *..:* ****::: :*:* : **
Mflv_3912|M.gilvum_PYR-GCK VGVYLDGGFGVGKTHLLASSYYEVAGRS--------DYRQAFATFGELTQ
Mvan_2491|M.vanbaalenii_PYR-1 VGVYLDGGFGVGKTHLLASSYYAVSGK------------KAFATFGELTQ
MSMEG_2788|M.smegmatis_MC2_155 VGLYLDGGFGVGKTHLLASTYYTLSQG---------NAKTAFATFGELTQ
TH_2402|M.thermoresistible__bu VGLYLDGGFGVGKTHLLASMYYTVPAP------------KAFATFGELTQ
Mb2689c|M.bovis_AF2122/97 VGLYLDGGFGVGKTHLLASAYYQLPGTG----PDAPTCPKAFATFGELTQ
Rv2670c|M.tuberculosis_H37Rv VGLYLDGGFGVGKTHLLASAYYQLPGTG----PDAPTCPKAFATFGELTQ
MMAR_2046|M.marinum_M VGLYLDGGFGVGKTHLLASSYYQLPGLG----PDAPEQPKAFATFGELTQ
MUL_3304|M.ulcerans_Agy99 VGLYLDGGFGVGNTHLLASSYYQLPGLG----PDAPEQPKAFATFGELTQ
MAV_3561|M.avium_104 VGLYLDGGFGVGKTHLLASAYYELPGDGPVRSPDGP-APKAFATFGELTQ
MAB_2975c|M.abscessus_ATCC_199 VGIYLDGGFGVGKTHLLASIYHTVPGP------------KAFASFGELTQ
**:*********:****** *: :. ***:******
Mflv_3912|M.gilvum_PYR-GCK LAGVFGFMECIELLSDYALVCIDEFELDDPGNTTLISRLLSALVERGVSV
Mvan_2491|M.vanbaalenii_PYR-1 LAGVFGFLECIDLLSDYVLVCIDEFELDDPGNTTLISRLLSALVERGVSV
MSMEG_2788|M.smegmatis_MC2_155 LAGVFGYNECIDLLAEYVVVCIDEFELDDPGNTTLISRLLSALVERGVSI
TH_2402|M.thermoresistible__bu LAGVFGFTECIDLLADYVLVCIDEFELDDPGNTTLISRLLSALVERGVSI
Mb2689c|M.bovis_AF2122/97 LAGVFGFADCIDLLANYTALCIDEFELDDPGNTTLISRLLSALVERGVSV
Rv2670c|M.tuberculosis_H37Rv LAGVFGFADCIDLLANYTALCIDEFELDDPGNTTLISRLLSALVERGVSV
MMAR_2046|M.marinum_M LAGVFGFAECIDLLANHTAVCIDEFELDDPGNTTLISRMLSALVQRGVSV
MUL_3304|M.ulcerans_Agy99 LAGVFGFAECIDLLANHTAVCIDEFELDDPGNTTLISRMLSALVQRGVSV
MAV_3561|M.avium_104 LAGVFGFAECVDLLADYTAVCIDEFELDDPGNTTLVARLLSSLVERGVSV
MAB_2975c|M.abscessus_ATCC_199 VASVFGFLECIELLADYTVVCIDEFELDDPGNTTLVSRLLSELVARGVSI
:*.***: :*::**:::. :***************::*:** ** ****:
Mflv_3912|M.gilvum_PYR-GCK AATSNTLPEQLGEGRFAAQDFLREINTLASMFTTVRIEGPDYRHRDLPPA
Mvan_2491|M.vanbaalenii_PYR-1 AATSNTLPEQLGEGRFAAQDFLREIHTLAAMFTTVRIEGPDYRHRDLPPA
MSMEG_2788|M.smegmatis_MC2_155 AATSNTLPEQLGEGRFAAQDFLREINTLAKIFTTVRIEGPDYRHRDLPPA
TH_2402|M.thermoresistible__bu AATSNTLPEQLGEGRFAAQDFLREINTLASIFTTIRIEGPDYRHRGLPPA
Mb2689c|M.bovis_AF2122/97 AATSNTLPEQLGEGRFAAQDFLREINTLASIFTTVRIEGPDYRHRDLPPA
Rv2670c|M.tuberculosis_H37Rv AATSNTLPEQLGEGRFAAQDFLREINTLASIFTTVRIEGPDYRHRDLPPA
MMAR_2046|M.marinum_M AATSNTLPEQLGEGRFAAQDFLREINTLASIFTTVRIEGPDYRHRGLPPA
MUL_3304|M.ulcerans_Agy99 AATSNTLPEQLGEGRFAAQDFLREINTLAGISTTVRIEGPDYRHRGLPPA
MAV_3561|M.avium_104 AATSNTLPEQLGEGRFAAQDFLREIKTLASIFTTVRIEGPDYRHRDLPPA
MAB_2975c|M.abscessus_ATCC_199 AATSNTLPEQLGEGRFAAQDFLREIAALSAIFNTVRVEGPDYRHRDLPPA
************************* :*: : .*:*:********.****
Mflv_3912|M.gilvum_PYR-GCK PEPPTDDEVARWAAEADGATLDDFDALCAHLATMHPSRYLTLIEGVSQVF
Mvan_2491|M.vanbaalenii_PYR-1 PEPPSDDDVARRAGEVSGATLDDFDALCAHLATMHPSRYLTLIEGVSQVF
MSMEG_2788|M.smegmatis_MC2_155 PEPLSDAEVAERAGGVDGATLDDFDALCAHLATMHPSRYHALIEGVSEVF
TH_2402|M.thermoresistible__bu PEPLSDAEVAARAAEVPGATLDDFDELCAHLASMHPSRYLTLIEGVRAVF
Mb2689c|M.bovis_AF2122/97 PAPLSDEEVAARAARVEGATLDDFDALCAHLATMHPSRYLTLIEGVTAVF
Rv2670c|M.tuberculosis_H37Rv PAPLSDEEVAARAARVEGATLDDFDALCAHLATMHPSRYLTLIEGVTAVF
MMAR_2046|M.marinum_M PQPLSDDQVAQRAATVRGATLDDFDALCAHLATMHPSRYLTLIDGVSAVF
MUL_3304|M.ulcerans_Agy99 PQPLSDDQVAQRAATVRGATLDDFDALCAHLATMHPSRYLTLIDGVSAVF
MAV_3561|M.avium_104 PPPLSDAQVAARAAVVPGATLDDFDALCAHLATMHPSRYLTLIEDVTAVF
MAB_2975c|M.abscessus_ATCC_199 PEPHSDEQLRELAGAINGATFDRFDDLCKHLASMHPSRYLTLIEGVTAVF
* * :* :: *. ***:* ** ** ***:****** :**:.* **
Mflv_3912|M.gilvum_PYR-GCK VTGVHPIDDQNVALRLVSLTDRLYDAGIPVVASGTKLDTIFSEEMLAGGF
Mvan_2491|M.vanbaalenii_PYR-1 VTGVHPIDDQNVALRLVSLTDRLYDAGIPVVASGTKLDTIFSQEMLEGGF
MSMEG_2788|M.smegmatis_MC2_155 ITGVHPIEDQSVALRLVALTDRLYDAGIPVLASGTKLDTIFSDEMLAGGF
TH_2402|M.thermoresistible__bu LTGVRPLDDQNVALRLVSLTDRLYDAGIPVVASGAKLDTIFSDEMLAGGF
Mb2689c|M.bovis_AF2122/97 LTGVHGIDDQNVALRLVALVDRLYDAGIPVVASGAKLDTIFSEEMLAGGY
Rv2670c|M.tuberculosis_H37Rv LTGVHGIDDQNVALRLVALVDRLYDAGIPVVASGAKLDTIFSEEMLAGGY
MMAR_2046|M.marinum_M LTGVHPIEDQHVALRLVALTDRLYDAGVPVVASGAKLDTIFSEEMLAGGY
MUL_3304|M.ulcerans_Agy99 LTGVHPIEDQHVALRLVALTDRLYDAGVPAVASGAKLDTIFSEEMLAGGY
MAV_3561|M.avium_104 LTGVHRIDDQNVALRLVSLTDRLYDAGVPVLASGEKLDTIFSAEMLSGGY
MAB_2975c|M.abscessus_ATCC_199 IADVEQVHDQSVALRIVALTDRLYDAGIPVQASGAKLNTIFDDEMVAGGF
::.*. :.** ****:*:*.*******:*. *** **:***. **: **:
Mflv_3912|M.gilvum_PYR-GCK RKKYLRATSRLLALTAAGQAGRTIT
Mvan_2491|M.vanbaalenii_PYR-1 RKKYLRATSRLLALTAAGQTERTIT
MSMEG_2788|M.smegmatis_MC2_155 RKKYLRATSRLLALTAAAQEKA---
TH_2402|M.thermoresistible__bu RKKYLRAMSRLLALTAAGHNGRIIT
Mb2689c|M.bovis_AF2122/97 RKKYLRATSRLLALTAGVIQAREP-
Rv2670c|M.tuberculosis_H37Rv RKKYLRATSRLLALTAGVIQAREP-
MMAR_2046|M.marinum_M RKKYLRATSRLLALTAPVSEAREP-
MUL_3304|M.ulcerans_Agy99 RKKYLRATSRLLALTAPVSEAREP-
MAV_3561|M.avium_104 RKKYLRAMSRLLALTAGANEAYGS-
MAB_2975c|M.abscessus_ATCC_199 RKKYLRATSRLLALSAAPLPNAA--
******* ******:*