For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAGDVARQRFWKDLRLNTQARIGLDRAGNALHTRDVLDLAAAHAIARDAVHVPLDIDRFAADVGALGLGV PVVVTSQVGSRAEYLRRPDLGRLPADLSAVARRPADIGFVLADGLSPTALMRHGVPLLRELALRLGNAYT LAAPVIATQARVALGDHVAEQGRYRTLIVIIGERPGLSVVDSLGIYLTHLPRSGCTDADRNCISNIHPPD GLSYSEAARIAASLVGGALQLGRSGVELKDTSRASAIDDPGLESLG*
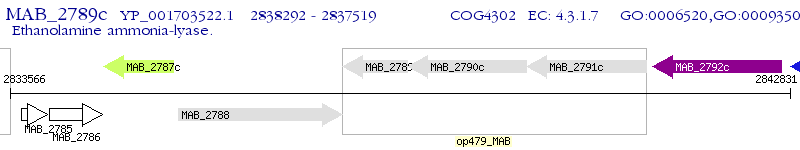
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2789c | - | - | 100% (257) | ethanolamine ammonia-lyase small subunit |
| M. abscessus ATCC 19977 | MAB_3827c | - | 5e-76 | 59.59% (245) | ethanolamine ammonia-lyase small subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0663 | eutC | 1e-82 | 65.75% (254) | ethanolamine ammonia-lyase light chain, EutC |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1554 | eutC | 1e-91 | 67.89% (246) | ethanolamine ammonia-lyase small subunit |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0109 | eutC | 1e-81 | 64.68% (252) | ethanolamine ammonia-lyase small subunit |
| M. vanbaalenii PYR-1 | Mvan_3198 | - | 3e-83 | 65.84% (243) | ethanolamine ammonia-lyase small subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0663|M.marinum_M MNAPDARAADEVWALLRDVTQARIGLGRAGNSLPTRRVLEFQAAHAAARD
MUL_0109|M.ulcerans_Agy99 MNAPDARAADEVWALLRDVTQARIGLGRAGNSLPTRRVLEFQAAHAAGRD
Mvan_3198|M.vanbaalenii_PYR-1 MSAADELPVEDVWAPLRRTTQARIGLGRAGNALPSQRVLEFKAAHAAARD
MSMEG_1554|M.smegmatis_MC2_155 MTVSNDIAVQQFWDELRKTTQARIGLGRAGNSLPTQQVLELAAAHAAARD
MAB_2789c|M.abscessus_ATCC_199 MTAG-DVARQRFWKDLRLNTQARIGLDRAGNALHTRDVLDLAAAHAIARD
*.. . : .* ** *******.****:* :: **:: **** .**
MMAR_0663|M.marinum_M AVHDPLDVAGLTEQVRDLGLGAPLVVRSQASSRSEYLRRPDLGRAPADLS
MUL_0109|M.ulcerans_Agy99 AVHDPLDVAGLTEQVRDLGLGAPLVVRSQASSRSEYLRRPELGRAPADLS
Mvan_3198|M.vanbaalenii_PYR-1 AVHVPLDAEALAIEVETLGLGAPAQVCSRAADRGEYLRRPDLGRAPADLS
MSMEG_1554|M.smegmatis_MC2_155 AVHVPLDVEALAAQVREVGIGEPIVVTSRATSRDEYLRRPDLGRQPAEGT
MAB_2789c|M.abscessus_ATCC_199 AVHVPLDIDRFAADVGALGLGVPVVVTSQVGSRAEYLRRPDLGRLPADLS
*** *** :: :* :*:* * * *:. .* ******:*** **: :
MMAR_0663|M.marinum_M ILPNTGADIGIVLADGLSPRALTDHGAGLLGALVREFHDRYRIAPPVIAT
MUL_0109|M.ulcerans_Agy99 ILPNTGADIGIVLADGLSPRALTDHGAGLLGALVREFHHRYRIAPPVIAT
Mvan_3198|M.vanbaalenii_PYR-1 ALPNSGADVGIVLADGLSPRALAEHGAGLLAALIDEFGGRYRLAPPVIAT
MSMEG_1554|M.smegmatis_MC2_155 EVPESGADIGFVLADGLSPMGLNHHGAALLKALVERLKDRHTLAPPIIAT
MAB_2789c|M.abscessus_ATCC_199 AVARRPADIGFVLADGLSPTALMRHGVPLLRELALRLGNAYTLAAPVIAT
:.. **:*:******** .* **. ** * .: : :*.*:***
MMAR_0663|M.marinum_M QARVALGDHVGQALGVRTLLVIIGERPGLSVADSLGIYLTHLPRPGLTDA
MUL_0109|M.ulcerans_Agy99 QARVALGDHIGQALGVRTLLVIIGERPGLSVADSLGVYLTHLPRPGLTDA
Mvan_3198|M.vanbaalenii_PYR-1 QARVALGDHIGAALGVSTLLMVIGERPGLSVADSLGIYLTHLPRPGRTDA
MSMEG_1554|M.smegmatis_MC2_155 QARVGLGDHIGAQARVRTLLVIIGERPGLSVADSLGIYLTHLPQPGRTDA
MAB_2789c|M.abscessus_ATCC_199 QARVALGDHVAEQGRYRTLIVIIGERPGLSVVDSLGIYLTHLPRSGCTDA
****.****:. **:::*********.****:******:.* ***
MMAR_0663|M.marinum_M DRNCISNIHPPDGLDYQTAAGIAAALVTGARKLGRSGVALKDTSR-TELT
MUL_0109|M.ulcerans_Agy99 DRNCISNIHPPDGLDYQTAAGIAAALVTGARKLGRSGVALKDTSR-TELT
Mvan_3198|M.vanbaalenii_PYR-1 DRNCISNIHPPDGLDYRRAARIAATLVAGARQLGRSGVALKDTSP-AELP
MSMEG_1554|M.smegmatis_MC2_155 DRNCISNIHPPDGLGYAEAARVASTLVDGAVALGRSGVDLKDTSRDAALG
MAB_2789c|M.abscessus_ATCC_199 DRNCISNIHPPDGLSYSEAARIAASLVGGALQLGRSGVELKDTSRASAID
**************.* ** :*::** ** ****** ***** : :
MMAR_0663|M.marinum_M ADDTLGIG
MUL_0109|M.ulcerans_Agy99 VDDTLGIG
Mvan_3198|M.vanbaalenii_PYR-1 G-------
MSMEG_1554|M.smegmatis_MC2_155 PLSPHELA
MAB_2789c|M.abscessus_ATCC_199 DPGLESLG