For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGLPAIPEQNRSPKSGYLFGAAAYLMWGLFPAFFPLLEPAGALEILAHRMIWTLLLMAVVVAVSRRLRDL VAMGLRNWGLLIGASLLIALNWGVYIYAVNSGHVLDAALGYYINPLVTVLLGVVLFREPLSRWQLLALVL AFTAVAVLTISVGRLPVITLILAASFAGYGAIKKTITVDPRVSLTAEGIVAAPFAGAYLVFVGVSGQQHF LGYTHGHTALMVLSGVVTALPLLLFGAAAQRIPLVTLGLLQYVTPTLQMLWGVLVKHEAMPPERWVGFVL IWAALVIFTADTVRSALILPRQSGRPATVSPIDMVSVKGRD*
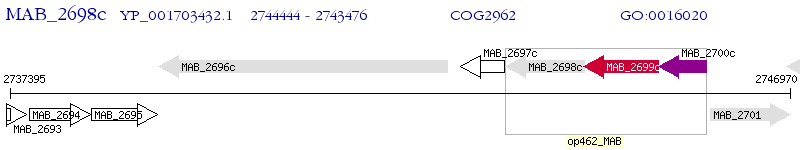
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2698c | - | - | 100% (322) | putative RarD protein |
| M. abscessus ATCC 19977 | MAB_3180 | - | 4e-05 | 23.98% (221) | hypothetical protein MAB_3180 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3638 | - | 2e-98 | 60.41% (293) | transporter DMT superfamily protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3176 | rarD | 9e-92 | 55.28% (284) | RarD protein |
| M. thermoresistible (build 8) | TH_0580 | rarD | 1e-86 | 53.74% (281) | RarD protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2775 | - | 2e-96 | 57.68% (293) | transporter DMT superfamily protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3638|M.gilvum_PYR-GCK -------------MRTGLFFGAGAYVMWGLFPAFFPLLKPAGALEILAHR
Mvan_2775|M.vanbaalenii_PYR-1 -------------MRSGLLYGAGAYVMWGLFPAFFPLLKPAGAVEILAHR
MAB_2698c|M.abscessus_ATCC_199 MSGLPAIPEQNRSPKSGYLFGAAAYLMWGLFPAFFPLLEPAGALEILAHR
MSMEG_3176|M.smegmatis_MC2_155 ---------MSGSNRTGVLYGAGAYIWWGLCPGFFLLLLPAGPLEVVAHR
TH_0580|M.thermoresistible__bu ---------MVNTRRQGLLCGVGAYLWWGLCPGYFPLLAPAGSVEVLAHR
: * : *..**: *** *.:* ** ***.:*::***
Mflv_3638|M.gilvum_PYR-GCK IIWSFVLMVVVVAAVRRMRDIAAISGRTWLLLTAASALIAANWLIYVYAV
Mvan_2775|M.vanbaalenii_PYR-1 VIWSFAMMVVVVAAVRRLGDIKAIDRRTWLLLVAAAALISANWLIFVYAV
MAB_2698c|M.abscessus_ATCC_199 MIWTLLLMAVVVAVSRRLRDLVAMGLRNWGLLIGASLLIALNWGVYIYAV
MSMEG_3176|M.smegmatis_MC2_155 IVWSTVFLMLVLAFARRLGDLRRLSLRTWLQLLAASALISINWGTYIFAV
TH_0580|M.thermoresistible__bu IVWSVVFLLVVLTVARRLGDLVRLSARTWLYLAAASALISLNWGTYIYAV
::*: :: :*:: **: *: :. *.* * .*: **: ** :::**
Mflv_3638|M.gilvum_PYR-GCK NNGHVVDAALGYFINPLVSIALGLVIFREKLNRWQGSALAIAVVAVVVLT
Mvan_2775|M.vanbaalenii_PYR-1 NNGHVVDAALGYFINPLVSIALGMLVFREKLNRWQFTALAIAVAAVAILT
MAB_2698c|M.abscessus_ATCC_199 NSGHVLDAALGYYINPLVTVLLGVVLFREPLSRWQLLALVLAFTAVAVLT
MSMEG_3176|M.smegmatis_MC2_155 TSGHVVDAALGYFINPLVSVALGVIIFRERINRWQAVSLALAVIAVAVLT
TH_0580|M.thermoresistible__bu MNDHVVDAALGYFINPLVSVALGVLVLGERVGGLQRIALAVAAVAVVVLT
..**:******:*****:: **:::: * :. * :*.:* **.:**
Mflv_3638|M.gilvum_PYR-GCK IQLGEPPVVGLGLALSFGLYGLVKKLVPTDPRVSVGIEAAIATPFAVAYL
Mvan_2775|M.vanbaalenii_PYR-1 VQLGQPPVVGLGLALSFGLYGAVKKVVRTDPRVSVGIEAAIATPFAIGYL
MAB_2698c|M.abscessus_ATCC_199 ISVGRLPVITLILAASFAGYGAIKKTITVDPRVSLTAEGIVAAPFAGAYL
MSMEG_3176|M.smegmatis_MC2_155 VEVGEPPYIAVVLAVSFGVYGLVKKVVQADPRVSVAVETLLALPVAGGYV
TH_0580|M.thermoresistible__bu VEVGQPPYIALVLAASFGIYGLVKKVVDADPRVGVAVETLLALPLAGGFL
:.:*. * : : ** **. ** :** : .****.: * :* *.* .::
Mflv_3638|M.gilvum_PYR-GCK VALQSGGGGTLTGHGGGHVALLILAGVLTALPLLLFARAAQLLPMVTLGL
Mvan_2775|M.vanbaalenii_PYR-1 VALQASGGGTLTGHGASHIALLILSGVLTALPLLLFAAAAQRLAMVSLGL
MAB_2698c|M.abscessus_ATCC_199 VFVGVSGQQHFLGYTHGHTALMVLSGVVTALPLLLFGAAAQRIPLVTLGL
MSMEG_3176|M.smegmatis_MC2_155 IVLEVLGRGHFIAESPWHPVLMVLSGVVTAVPLLLFAAAAQRLTMVMMGL
TH_0580|M.thermoresistible__bu IALHLLGEGSFGNHGAGHTVLLLAAGPVTAIPLLLFAAAAQRLPLVVLGL
: : * : * .*:: :* :**:*****. *** :.:* :**
Mflv_3638|M.gilvum_PYR-GCK LFYVTPTMQLTWGVLVGGEPMPPMRWFGFALIWVALAVFTVDAVRRARAD
Mvan_2775|M.vanbaalenii_PYR-1 LFYVNPAMQLSWGVLVGGEPMPPMRWLGFALIWLALLVFTGDALRRARVQ
MAB_2698c|M.abscessus_ATCC_199 LQYVTPTLQMLWGVLVKHEAMPPERWVGFVLIWAALVIFTADTVRSALIL
MSMEG_3176|M.smegmatis_MC2_155 LFYLNPALQMLWGVFIGHEPMPLGRWIGFALIWTALAVMTIGTLRNRRS-
TH_0580|M.thermoresistible__bu LFYLNPALQMAWGVLVNHESMPLGRWIGFALIWVALAVMAVDALRRGGAG
* *:.*::*: ***:: *.** **.**.*** ** ::: .::*
Mflv_3638|M.gilvum_PYR-GCK RRVSATPVA-------------
Mvan_2775|M.vanbaalenii_PYR-1 RRAARPCPA-------------
MAB_2698c|M.abscessus_ATCC_199 PRQSGRPATVSPIDMVSVKGRD
MSMEG_3176|M.smegmatis_MC2_155 DAVADAAQTQPTLP--------
TH_0580|M.thermoresistible__bu TPNRPAAVPDRVVP--------
.