For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FVGGHPTVNPPRTFGGQLMAQSLVASGRTVSEKLSLAAISTHFVNGGDPELDIEFRVQRLRDERNFANRR VDAVQGDLLLSSALVSYLKDSPGLEHAVNPPVVADPETLPSIDEHLVGYEETVPMFVAALKPIEWRYDND PAWVLKAKSQRLEHNRVWMKTHAALPDDPVLHSAALVYSSDTTILDSIITTHGLSWGYDRIFAATINHSV WFHRPLRFDDWVLYATESPVASAGRGMGAGRYFDRDGALLASTIQEGIVKYFPGRS*
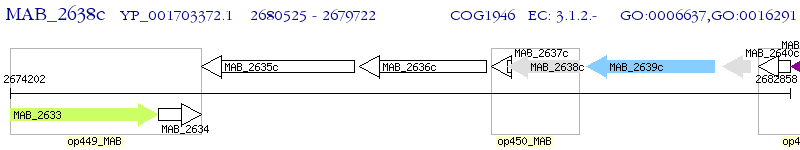
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2638c | - | - | 100% (267) | acyl-CoA thioesterase |
| M. abscessus ATCC 19977 | MAB_2891c | - | 7e-41 | 36.26% (262) | acyl-CoA thioesterase II TesB2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1644 | tesB1 | 1e-104 | 66.04% (265) | acyl-CoA thioesterase II TesB1 |
| M. gilvum PYR-GCK | Mflv_3590 | - | 1e-101 | 63.50% (263) | acyl-CoA thioesterase |
| M. tuberculosis H37Rv | Rv1618 | tesB1 | 1e-103 | 65.66% (265) | acyl-CoA thioesterase II TesB1 |
| M. leprae Br4923 | MLBr_01278 | tesB | 1e-100 | 62.64% (265) | acyl CoA thioesterase II |
| M. marinum M | MMAR_2421 | tesB1 | 1e-102 | 64.39% (264) | acyl-CoA thioesterase II TesB1 |
| M. avium 104 | MAV_3169 | - | 1e-105 | 66.79% (265) | Acyl-CoA thioesterase |
| M. smegmatis MC2 155 | MSMEG_3228 | - | 1e-108 | 66.17% (266) | acyl-CoA thioesterase II |
| M. thermoresistible (build 8) | TH_4001 | - | 1e-111 | 70.57% (265) | PUTATIVE acyl-CoA thioesterase |
| M. ulcerans Agy99 | MUL_1599 | tesB1 | 1e-102 | 64.02% (264) | acyl-CoA thioesterase II TesB1 |
| M. vanbaalenii PYR-1 | Mvan_2825 | - | 1e-105 | 65.28% (265) | acyl-CoA thioesterase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2421|M.marinum_M MPAGDEPNVRDEAKSDFEELLAVLDLKRVADDRFIGAHPSKNPMRTFGGL
MUL_1599|M.ulcerans_Agy99 --------MRDEAKSDFEELLAVLDLKRVADDRFIGAHPSKNPMRTFGGL
MAV_3169|M.avium_104 MPAGAE-SQEPNTTADFEELLATLDLRRVADDLFVGSHPSKNPMRTFGGQ
Mb1644|M.bovis_AF2122/97 -------MPDGKPMSDFDELLAVLDLNAVASDLFTGSHPSKNPLRTFGGQ
Rv1618|M.tuberculosis_H37Rv -------MPDGKPMSDFDELLAVLDLNAVASDLFTGSHPSKNPLRTFGGQ
MLBr_01278|M.leprae_Br4923 ----MKRLWEKKTMSDFEELLAVLDLNRVADDLFIGSHPSKNPLRTFGGQ
Mflv_3590|M.gilvum_PYR-GCK ----------MSRHADFDELLAILHLDRVDENTFIGRHPSKNPVRTFGGQ
Mvan_2825|M.vanbaalenii_PYR-1 ----------MSSQADFDELLAVLDLDRLDDDTFVGSHPSKNPVRTFGGQ
MSMEG_3228|M.smegmatis_MC2_155 ----------MS-QADFEELLAVLDLERVGEDLFIGSHPSKNPVRTFGGQ
TH_4001|M.thermoresistible__bu ----------LAAASDFEELLAILDLRRTDDSTFLGSHPSKNPVRTFGGQ
MAB_2638c|M.abscessus_ATCC_199 --------------------------------MFVGGHPTVNPPRTFGGQ
* * **: ** *****
MMAR_2421|M.marinum_M LMAQSFVASSRTLLRDDLPPSALSVHFINGGDTTKDIEFQVVRLRDERRF
MUL_1599|M.ulcerans_Agy99 LMAQSFVANSRTLLRDDLPPSALSVHFINGGDTTKDIEFQVVRLRDERRF
MAV_3169|M.avium_104 LMSQSFVASSRSLIRDDLPPSALSVHFINGGDTGKDIEYRVTRLRDERRF
Mb1644|M.bovis_AF2122/97 LMAQSFVASSRTLTRHHLPPSAFSVHFINGGDTAKDIEFQVIRLRDERRF
Rv1618|M.tuberculosis_H37Rv LMAQSFVASSRTLTRHHLPPSAFSVHFINGGDTAKDIEFQVIRLRDERRF
MLBr_01278|M.leprae_Br4923 LVAQSFVASSRTLIRDDLPPSAFSVHFINGGDTAKEIEFHVVRLRDERRF
Mflv_3590|M.gilvum_PYR-GCK MMAQAFVAAGRSLN-HPTPPSALSAHFIAGGDPEQDLEFHVVRLRDERRF
Mvan_2825|M.vanbaalenii_PYR-1 MMAQAFVAASRSLR-HPTPPSALSVHFIAGGDPALDLEFHVVRLRDERRF
MSMEG_3228|M.smegmatis_MC2_155 MTAQAFVAAGRSLE-HKLSPSALSAHFIAGGDPEKDLEFHIVRLRDERRF
TH_4001|M.thermoresistible__bu MMAQAFVAAGRTVP-AQLPPSAISVHFIAGGDPARDLEFRVVRLRDERRF
MAB_2638c|M.abscessus_ATCC_199 LMAQSLVASGRTVS-EKLSLAAISTHFVNGGDPELDIEFRVQRLRDERNF
: :*::** .*:: . :*:*.**: ***. ::*::: ******.*
MMAR_2421|M.marinum_M ANRRIDAVQNGTLLSSAMISYMSGGRGLEHGVEPPPVADPETRPPIGELL
MUL_1599|M.ulcerans_Agy99 ANRRIDAVQNGTLLSSAMISYMSGGRGLEHGVEPPPVADPETRPPIGELL
MAV_3169|M.avium_104 ANRRVDAVQDGTLLCSALISYMAGGRGLEHGVDPPRVADPETQPTIVELL
Mb1644|M.bovis_AF2122/97 ANRRVDAVQDGTLLSSAMVSYMAGGRGLEHALDPPQVAEPHTRPPIGELL
Rv1618|M.tuberculosis_H37Rv ANRRVDAVQDGTLLSSAMVSYMAGGRGHEHALDPPQVAEPHTRPPIGELL
MLBr_01278|M.leprae_Br4923 ANRRVDAIQDGMLLSSAMASYMSGGCGLEHAVEPPEAAEPHTRPPIGELL
Mflv_3590|M.gilvum_PYR-GCK ANRRVDVMQNGQLLTTVLVSYLAGGKGLEHGVEPPQIDDPEAVLPIDELL
Mvan_2825|M.vanbaalenii_PYR-1 ANRRVDVMQKGQLLTTAMVSYLSGGKGLEHGVEPPPLDDPLTVPPIDDLL
MSMEG_3228|M.smegmatis_MC2_155 ANRRVDVMQNGELLTTVMISYMNGGRGLEHNVPAPDVPRPETLPKIDELL
TH_4001|M.thermoresistible__bu ANRRVDVCQGDDLLATAMVSYLSGGRGLEHSVAPPDVPGPEELPKIDELL
MAB_2638c|M.abscessus_ATCC_199 ANRRVDAVQGDLLLSSALVSYLKDSPGLEHAVNPPVVADPETLPSIDEHL
****:*. * . ** :.: **: .. * ** : .* * * : *
MMAR_2421|M.marinum_M RGYEETVPHFVNALQPIEWRYTNDPSWVMRDKGERLHYNRVWVKALGKLP
MUL_1599|M.ulcerans_Agy99 RGYEETVPHFVNALQPIEWRYTNDPSWVMRDKGERLHYNRVWVKALGKLP
MAV_3169|M.avium_104 RGYEETVPHFVNALQPIEWRYTNDPAWVMRTKGERLPHNRVWVKALGKLP
Mb1644|M.bovis_AF2122/97 RGYEETVPHFVNALQPIEWRYANDPAWIMRDKGDRLAYNRVWVKALGEMP
Rv1618|M.tuberculosis_H37Rv RGYEETVPHFVNALQPIEWRYANDPAWIMRDKGDRLAYNRVWVKALGEMP
MLBr_01278|M.leprae_Br4923 RGYEQIVQHFVNALQPVEWRYVNDPSWVMRNKGQRLAYNRVWVKALGALP
Mflv_3590|M.gilvum_PYR-GCK RGYEKIVPHFADALRPIEWRYTNDPAWIMRDKGGTLGHNRVWMTAQGPMP
Mvan_2825|M.vanbaalenii_PYR-1 RGYEQVVPHFANASRPIEWRYTNDPAWIMRDKGEKLTHNRVWMKAHGALP
MSMEG_3228|M.smegmatis_MC2_155 RGYEETVPLFVNALRPIEWRYTNDPAWVMRDKGGKLDHNRVWLKTEGEMP
TH_4001|M.thermoresistible__bu RGYEETVPHFVEALRPIEWRYANDPAWVMRDKGGRLEYNRVWLKTEGALP
MAB_2638c|M.abscessus_ATCC_199 VGYEETVPMFVAALKPIEWRYDNDPAWVLKAKSQRLEHNRVWMKTHAALP
***: * *. * :*:**** ***:*::: *. * :****:.: . :*
MMAR_2421|M.marinum_M DDPILHTATMLYSSDTTVLDSVITTHGLSWGFDRIFAASANHSVWFHRQV
MUL_1599|M.ulcerans_Agy99 DDPILHTATMLYSSDTTVLDSVITTHGLSWGFDRIFAASANHSVWFHRQV
MAV_3169|M.avium_104 DDPVLHTAAMVYSSDTTVLDSVITTHGLSWGFDRIFAASANHSIWFHRQV
Mb1644|M.bovis_AF2122/97 DDPVLHTATLLYSSDTTVLDSVITTHGLSWGFDRIFAASANHSVWFHRQV
Rv1618|M.tuberculosis_H37Rv DDPVLHTATLLYSSDTTVLDSVITTHGLSWGFDRIFAASANHSVWFHRQV
MLBr_01278|M.leprae_Br4923 NDPTLHTAAMLYSTDTTVLDSVITTHGLSWGFDRIFAVSANHSVWFHRQV
Mflv_3590|M.gilvum_PYR-GCK DDQVLHGAALVYSSDTTVLDSIITTHGLSWGHDRIFAVTTNHTVWFHRPI
Mvan_2825|M.vanbaalenii_PYR-1 DDPVLHTAAMVYSSDTTVLDSIITTHGLSWGHDRIFAVTTNHSVWFHRQV
MSMEG_3228|M.smegmatis_MC2_155 DDPVLHGAALVYSSDTTVLDSIITTHGLSWGFDRIFAVTMNHSVWFHRPI
TH_4001|M.thermoresistible__bu DDPVLHAAAMLYSSDTTVLDSIITTHGLSWGFDRIFAVTTNHSVWFHRPV
MAB_2638c|M.abscessus_ATCC_199 DDPVLHSAALVYSSDTTILDSIITTHGLSWGYDRIFAATINHSVWFHRPL
:* ** *:::**:***:***:*********.*****.: **::**** :
MMAR_2421|M.marinum_M DFNDWVLYSTSSPVAADSRGLGTGHFFNRTGQPVATVVQEGVLKYFPAAR
MUL_1599|M.ulcerans_Agy99 DFNDWVLYSTSSPVAADSRGLGTGHFFNRTGQPVATVVQEGVLKYFPAAR
MAV_3169|M.avium_104 DFNDWVLYSTSSPVAADSRGLGTGHFFDRSGQPLATVMQEGVLKYFPSAR
Mb1644|M.bovis_AF2122/97 NFDDWVLYSTSSPVAADSRGLGSGHFFDRSGKLIATVVQEGVLKYFPATP
Rv1618|M.tuberculosis_H37Rv NFDDWVLYSTSSPVAADSRGLGSGHFFDRSGKLIATVVQEGVLKYFPATP
MLBr_01278|M.leprae_Br4923 NFDDWVLYSTSSPVAADSRGLGTGHLFDRSGQLIATVVQEGVLKYFPASH
Mflv_3590|M.gilvum_PYR-GCK RFDEWVLYSTTSPVAADSRGLGTGHFFDRSGQPLATVVQEGIVKHFP---
Mvan_2825|M.vanbaalenii_PYR-1 RFDDWVLYSTTSPVAADSRGLGTGHFFDRSGLPLATVVQEGIVKHFPGRV
MSMEG_3228|M.smegmatis_MC2_155 RFDEWVLYSTTSPVAAESRGLGTGHYFDASGQLVATVVQEGIVKYFPGK-
TH_4001|M.thermoresistible__bu RFDEWVLYSTTSPVAAESRGLGSGHFFDRSGQLLATVIQEGIVKYFPAR-
MAB_2638c|M.abscessus_ATCC_199 RFDDWVLYATESPVASAGRGMGAGRYFDRDGALLASTIQEGIVKYFPGRS
*::****:* ****: .**:*:*: *: * :*:.:***::*:**
MMAR_2421|M.marinum_M R------
MUL_1599|M.ulcerans_Agy99 R------
MAV_3169|M.avium_104 R------
Mb1644|M.bovis_AF2122/97 DSAAGRS
Rv1618|M.tuberculosis_H37Rv DSAAGRS
MLBr_01278|M.leprae_Br4923 R------
Mflv_3590|M.gilvum_PYR-GCK -------
Mvan_2825|M.vanbaalenii_PYR-1 TASG---
MSMEG_3228|M.smegmatis_MC2_155 -------
TH_4001|M.thermoresistible__bu -------
MAB_2638c|M.abscessus_ATCC_199 -------