For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MASDRFDPHALITESRLGILATIKSSGLPQLSPVTPYYDRDNGIIYVSMTDGRAKTANLRRDPRAALECT SSDGRAWATAEGHVTLTGPGADPDGPEVQALVDYYRGAAGEHPDWDEYRSVMVSDRRVLMALRVERVYGQ KLG
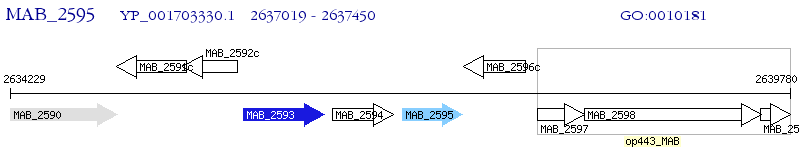
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2595 | - | - | 100% (143) | putative pyridoxamine 5'-phosphate oxidase |
| M. abscessus ATCC 19977 | MAB_2542 | - | 2e-10 | 28.46% (130) | hypothetical protein MAB_2542 |
| M. abscessus ATCC 19977 | MAB_2193c | - | 6e-06 | 27.82% (133) | hypothetical protein MAB_2193c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1186 | - | 4e-34 | 51.06% (141) | hypothetical protein Mb1186 |
| M. gilvum PYR-GCK | Mflv_2137 | - | 2e-36 | 53.10% (145) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
| M. tuberculosis H37Rv | Rv1155 | - | 3e-33 | 50.35% (141) | hypothetical protein Rv1155 |
| M. leprae Br4923 | MLBr_01508 | - | 2e-31 | 48.94% (141) | hypothetical protein MLBr_01508 |
| M. marinum M | MMAR_4298 | - | 2e-35 | 53.85% (130) | hypothetical protein MMAR_4298 |
| M. avium 104 | MAV_1292 | - | 2e-35 | 53.73% (134) | pyridoxamine 5'-phosphate oxidase family protein |
| M. smegmatis MC2 155 | MSMEG_6576 | - | 6e-63 | 79.43% (141) | pyridoxamine 5'-phosphate oxidase family protein |
| M. thermoresistible (build 8) | TH_0458 | - | 2e-36 | 54.48% (134) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0996 | - | 4e-36 | 54.62% (130) | hypothetical protein MUL_0996 |
| M. vanbaalenii PYR-1 | Mvan_4567 | - | 6e-35 | 51.03% (145) | pyridoxamine 5'-phosphate oxidase-related, FMN-binding |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4298|M.marinum_M ----------------MGRQVFDDKLLAVISGNSIGVLATIKHDGRPQLS
MUL_0996|M.ulcerans_Agy99 ----------------MGRQVFDDKLLAVISGNSIGVLATIKHDGRPQLS
MLBr_01508|M.leprae_Br4923 MRFGASLLRVRRRLYGMGSQVFDDKLLAVISGNSIGVLATIKRDGRPQLS
Mb1186|M.bovis_AF2122/97 ----------------MARQVFDDKLLAVISGNSIGVLATIKHDGRPQLS
Rv1155|M.tuberculosis_H37Rv ----------------MARQVFDDKLLAVISGNSIGVLATIKHDGRPQLS
MAV_1292|M.avium_104 ----------------MGRQVFDDKLLAVISGNSLGVLATIKRDGRPQLS
TH_0458|M.thermoresistible__bu ----------------MTRRVFDDKLLAVIAGNSLGVLATIKRDGRPQLS
Mflv_2137|M.gilvum_PYR-GCK ----------------MTRDVFDDKLLALLAGNSLGVLATLKRDGRPQLS
Mvan_4567|M.vanbaalenii_PYR-1 ----------------MTRDVFDDKLLALIGGNSLGVLATLKRDGRPQLS
MAB_2595|M.abscessus_ATCC_1997 ----------------MASDRFDPH--ALITESRLGILATIKSSGLPQLS
MSMEG_6576|M.smegmatis_MC2_155 -------------MTATSSDAFDPR--SLLAEARIGVLATIKANGLPQLS
** : ::: :*:***:* .* ****
MMAR_4298|M.marinum_M NVQYYFDARKLVVQVSITEPRAKTRNLRRDPRASILVDADDGWSYAVAEG
MUL_0996|M.ulcerans_Agy99 NVQYYFDARKLVVQVSITEPRAKTRNLRRDPRASILVDADDGRSYAVAEG
MLBr_01508|M.leprae_Br4923 NVQYHFDPRDLAVRVSITEPGVKTRNLRRDPRASILVDVDDGWSYAVAEG
Mb1186|M.bovis_AF2122/97 NVQYHFDPRKLLIQVSIAEPRAKTRNLRRDPRASILVDADDGWSYAVAEG
Rv1155|M.tuberculosis_H37Rv NVQYHFDPRKLLIQVSIAEPRAKTRNLRRDPRASILVDADDGWSYAVAEG
MAV_1292|M.avium_104 NVQYHFDPRSLVIQVSITEPRAKTRNLRRDPRASILVDADDGWSYAVAEG
TH_0458|M.thermoresistible__bu NVTYYFDPRRLTIQVSVTEPRAKTRNLRRDPRASIHVSSDDGWSYAVAEG
Mflv_2137|M.gilvum_PYR-GCK NVSYFFDPRALTISVSITEPRAKTRNLRRDPRAAIHVSSDDGWAYAVAEG
Mvan_4567|M.vanbaalenii_PYR-1 NVSYHFDARTLTISVSIAEPRAKTRNLRRDPRAAIHVSSDDGWAYAVAEG
MAB_2595|M.abscessus_ATCC_1997 PVTPYYDRDNGIIYVSMTDGRAKTANLRRDPRAALECTSSDGRAWATAEG
MSMEG_6576|M.smegmatis_MC2_155 PVTPYYDRAADIIYVSMTDGRAKTANLRRDPRAALEVTRADGWAWATAEG
* .:* : **::: .** ********:: ** ::*.***
MMAR_4298|M.marinum_M TAELTPPAAAPDDETVEALIALYRNIAGEHPDWDEYREAMVTDRRVLLTL
MUL_0996|M.ulcerans_Agy99 TAELTPPAAAPDDETVEALIALYRNIAGEHPDWDEYREAMVIDRRVLLTL
MLBr_01508|M.leprae_Br4923 TAELTPPAAAPDDDTVEALIVLYRNIVGEHLDWDEYRQAMVTDRRVLLTL
Mb1186|M.bovis_AF2122/97 TAQLTPPAAAPDDDTVEALIALYRNIAGEHPDWDDYRQAMVTDRRVLLTL
Rv1155|M.tuberculosis_H37Rv TAQLTPPAAAPDDDTVEALIALYRNIAGEHSDWDDYRQAMVTDRRVLLTL
MAV_1292|M.avium_104 TAELTPPAAAPDDDTVQGLIALYRNIAGEHPDWDEYRQAMVTDRRVLLTL
TH_0458|M.thermoresistible__bu TVTLTPEAADPYDDTVEGLIELYRNIAGEHPDWDDYRRAMVEDRRVLLTM
Mflv_2137|M.gilvum_PYR-GCK DAVLSPPAANPDDDTVAGLVELYRSLAGEHPDWDDYRRAMVDDRRVLMTL
Mvan_4567|M.vanbaalenii_PYR-1 DAVLSAPAADPGDDTVTGLIELYRNISGEHPDWDDYRRAMVDERRVLMRL
MAB_2595|M.abscessus_ATCC_1997 HVTLTGPGADPDGPEVQALVDYYRGAAGEHPDWDEYRSVMVSDRRVLMAL
MSMEG_6576|M.smegmatis_MC2_155 TVTLTGPGTDPHGPEVEALVEYYRKAAGEHPDWDEYRSVMVSDRRVLMKL
. *: .: * . * .*: ** *** ***:** .** :****: :
MMAR_4298|M.marinum_M PISHVYGLPPGKR
MUL_0996|M.ulcerans_Agy99 PISHVYGLPPGKR
MLBr_01508|M.leprae_Br4923 PILHVYGLPPGIR
Mb1186|M.bovis_AF2122/97 PISHVYGLPPGMR
Rv1155|M.tuberculosis_H37Rv PISHVYGLPPGMR
MAV_1292|M.avium_104 PISHVYGMPPGMR
TH_0458|M.thermoresistible__bu PISHVYGMPPGMR
Mflv_2137|M.gilvum_PYR-GCK PISHVYGMPPGKR
Mvan_4567|M.vanbaalenii_PYR-1 PISHVYGMPPGKR
MAB_2595|M.abscessus_ATCC_1997 RVERVYGQKLG--
MSMEG_6576|M.smegmatis_MC2_155 SVQRVYGERLR--
: :***