For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIGIVALAIGVVLGLVFHPNVPDAVAPYLPIAVVAALDALFGGARAYLDQIFDSKVFVVSFVFNVLVAAL IVFVGDQLGVGTQLSTAIIVVLGIRIFGNAAALRRRLFGA
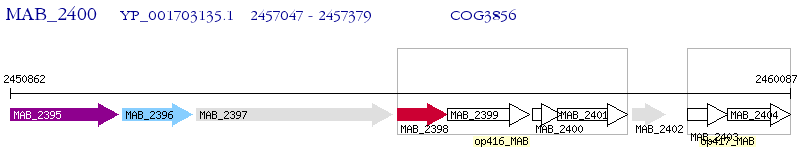
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2400 | - | - | 100% (110) | hypothetical protein MAB_2400 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1855 | - | 1e-49 | 86.36% (110) | hypothetical protein Mb1855 |
| M. gilvum PYR-GCK | Mflv_3405 | - | 1e-48 | 85.45% (110) | hypothetical protein Mflv_3405 |
| M. tuberculosis H37Rv | Rv1824 | - | 1e-49 | 86.36% (110) | hypothetical protein Rv1824 |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2701 | - | 1e-50 | 88.18% (110) | hypothetical protein MMAR_2701 |
| M. avium 104 | MAV_2891 | - | 7e-50 | 87.27% (110) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_3650 | - | 2e-49 | 87.27% (110) | integral membrane protein |
| M. thermoresistible (build 8) | TH_1123 | - | 3e-48 | 86.36% (110) | CONSERVED HYPOTHETICAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_3043 | - | 1e-50 | 88.18% (110) | hypothetical protein MUL_3043 |
| M. vanbaalenii PYR-1 | Mvan_3138 | - | 2e-49 | 86.36% (110) | hypothetical protein Mvan_3138 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3405|M.gilvum_PYR-GCK ------------MIGILALAVGIVVGVVFQPSVPEFVQPYLPIAVVAALD
Mvan_3138|M.vanbaalenii_PYR-1 -----MFDGCPLLIGIVALVVGIVLGVVFHPDVPEFAQPYLPIAVVAALD
MSMEG_3650|M.smegmatis_MC2_155 ------------MIGIVALVVGIVLGLVFRPDVPEVVEPYLPIAVVAALD
TH_1123|M.thermoresistible__bu ------------MIGIAALVVGIVLGLVFQPDVPVLVQPYLPIAVVAALD
Mb1855|M.bovis_AF2122/97 -MGSDTAWSPARMIGIAALAVGIVLGLVFHPGVPEVIQPYLPIAVVAALD
Rv1824|M.tuberculosis_H37Rv -MGSDTAWSPARMIGIAALAVGIVLGLVFHPGVPEVIQPYLPIAVVAALD
MAV_2891|M.avium_104 ------------MIGIAALAIGIVLGLVFHPSVPEVVQPYLPIAVVAALD
MMAR_2701|M.marinum_M ------------MIGIAALAVGIVLGLVFHPSVPEAIQPYLPIAVVAALD
MUL_3043|M.ulcerans_Agy99 MRGREADGKSASMIGIAALAVGIVLGLVFHPSVPEAIQPYLPIAVVAALD
MAB_2400|M.abscessus_ATCC_1997 ------------MIGIVALAIGVVLGLVFHPNVPDAVAPYLPIAVVAALD
:*** **.:*:*:*:**:*.** ************
Mflv_3405|M.gilvum_PYR-GCK AVFGGLRAYLENIFDSKVFVVSFVFNVLVAALIVYVGDQLGVGTQLSTAI
Mvan_3138|M.vanbaalenii_PYR-1 AVFGGLRAYLESIFDSKVFVVSFVFNVLVAALIVYVGDQLGVGTQLSTAI
MSMEG_3650|M.smegmatis_MC2_155 AVFGGLRAYLEKIFDSKVFVVSFVFNVLVAALIVYLGDQLGVGTQLSTAI
TH_1123|M.thermoresistible__bu AVFGGLRAYLEGIFDSKVFVVSFVFNVLVAALIVYLGDQLGVGTQLSTAI
Mb1855|M.bovis_AF2122/97 AVFGGLRAYLERIFDPKVFVVSFVFNVLVAALIVYVGDQLGVGTQLSTAI
Rv1824|M.tuberculosis_H37Rv AVFGGLRAYLERIFDPKVFVVSFVFNVLVAALIVYVGDQLGVGTQLSTAI
MAV_2891|M.avium_104 AVFGGLRAYLERIFDPKVFVISFVFNVFVAALIVYVGDQLGVGTQLSTAI
MMAR_2701|M.marinum_M AVFGGLRAYLERIFDPKVFVVSFVFNVLVAALIVYVGDQLGVGTQLSTAI
MUL_3043|M.ulcerans_Agy99 AVFGGLRAYLERIFDPKVFVVSFVFNVLVAALIVYVGDQLGVGTQLSTAI
MAB_2400|M.abscessus_ATCC_1997 ALFGGARAYLDQIFDSKVFVVSFVFNVLVAALIVFVGDQLGVGTQLSTAI
*:*** ****: ***.****:******:******::**************
Mflv_3405|M.gilvum_PYR-GCK IVVLGIRIFGNAAALRRRLFGA
Mvan_3138|M.vanbaalenii_PYR-1 IVVLGIRIFGNAAALRRRLFGA
MSMEG_3650|M.smegmatis_MC2_155 IVVLGIRIFGNAAALRRRLFGA
TH_1123|M.thermoresistible__bu IVVLGIRIFGNAAALRRRLFGA
Mb1855|M.bovis_AF2122/97 IVVLGIRIFGNTAALRRRLFGA
Rv1824|M.tuberculosis_H37Rv IVVLGIRIFGNTAALRRRLFGA
MAV_2891|M.avium_104 IVVLGIRIFGNAAALRRRLFGA
MMAR_2701|M.marinum_M IVVLGIRIFGNAAALRRRLFGA
MUL_3043|M.ulcerans_Agy99 IVVLGIRIFGNAAALRRRLFGA
MAB_2400|M.abscessus_ATCC_1997 IVVLGIRIFGNAAALRRRLFGA
***********:**********