For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSSDGSAERKTLLVFADSLSYYGPTGGLAASDPRIWPNIVAKKLDWDLELIARIGWTCRDVWWAAIQDP RAWAAVPTAGAVIFATGGMDSLPSPLPTALRESIRLIRPAKLRSWVREGYGWLQPRLSPIARVALPPKLT VEYLEKTRGALDFNRPGLPMVASLPSVHIAESYGRVHSGREATASAIAAWASEHKIPVVDLKQGVGEEVF SGRANPDGIHWNFVAHERVAELMIDALQSVLPELKAR
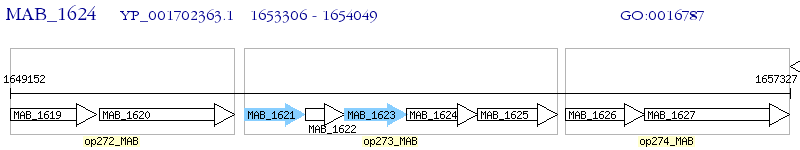
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1624 | - | - | 100% (247) | hypothetical protein MAB_1624 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2441c | - | 1e-104 | 71.43% (245) | hypothetical protein Mb2441c |
| M. gilvum PYR-GCK | Mflv_2656 | - | 1e-105 | 74.04% (235) | hypothetical protein Mflv_2656 |
| M. tuberculosis H37Rv | Rv2418c | - | 1e-104 | 71.43% (245) | hypothetical protein Rv2418c |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3746 | - | 1e-102 | 70.00% (250) | hypothetical protein MMAR_3746 |
| M. avium 104 | MAV_1753 | - | 1e-108 | 75.97% (233) | hypothetical protein MAV_1753 |
| M. smegmatis MC2 155 | MSMEG_4578 | - | 1e-106 | 73.73% (236) | hypothetical protein MSMEG_4578 |
| M. thermoresistible (build 8) | TH_0786 | - | 1e-103 | 71.31% (237) | HYPOTHETICAL PROTEIN Rv2418c |
| M. ulcerans Agy99 | MUL_3690 | - | 1e-102 | 69.60% (250) | hypothetical protein MUL_3690 |
| M. vanbaalenii PYR-1 | Mvan_3923 | - | 1e-102 | 68.70% (246) | hypothetical protein Mvan_3923 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4578|M.smegmatis_MC2_155 MSSETSSESTGHRPVLLVFADSLSYFGPTGGLPADDPRIWPNIVGEQLGW
TH_0786|M.thermoresistible__bu MSSDRGD---GRPPVLLVFADSLAYYGPTGGLPADDPRIWPNLVAQQLGW
Mflv_2656|M.gilvum_PYR-GCK MSSDPT----GVRRTLLIFCDSLSYYGPTGGLPSDDPRIWPNLVAGQLDW
Mvan_3923|M.vanbaalenii_PYR-1 MSSEAAR---GGARTLLVFCDSLSYYGPTGGLPSDDPRIWPNIVAAELGW
Mb2441c|M.bovis_AF2122/97 MSSRRGR-----RPALLVFADSLAYYGPTGGLPADDPRIWPNIVASQLDW
Rv2418c|M.tuberculosis_H37Rv MSSRRGR-----RPALLVFADSLAYYGPTGGLPADDPRIWPNIVASQLDW
MMAR_3746|M.marinum_M MSSERGASP-SQRPLLLVFADSLSYYGPTGGLPADDPRIWPNIVAAQLGW
MUL_3690|M.ulcerans_Agy99 MSSERGASP-SQRPLLLVFADSLSYYGPTGGLPADDPRIWPNIVAAQLGW
MAV_1753|M.avium_104 MSSEQRP-----RPTLLIFADSLAYYGPTGGLPADDPRIWPNIVAAQLDW
MAB_1624|M.abscessus_ATCC_1997 MTSSDGS---AERKTLLVFADSLSYYGPTGGLAASDPRIWPNIVAKKLDW
*:* **:*.***:*:******.:.*******:*. :*.*
MSMEG_4578|M.smegmatis_MC2_155 DVELIGRIGWTCRDVWWAATQDPRSWAALPRAGAVVFATSGMDSLPSPLP
TH_0786|M.thermoresistible__bu DVELIGRIGWTSRDVWWAATQDPRAWAALPRAGAVIFATGGMDSLPSPLP
Mflv_2656|M.gilvum_PYR-GCK DVELIGRIGWTSRDVWWAATQDPRSWAALPRAGAVIFATSGMDSLPSPLP
Mvan_3923|M.vanbaalenii_PYR-1 DVELIGRIGWTSRDVWWAATQDPRSWAALPRAGAVIFATSGMDSLPSPLP
Mb2441c|M.bovis_AF2122/97 DLELIGRIGWTCRDVWWAATQDPRAWAALPRAGAVIFATGGMDSLPSVLP
Rv2418c|M.tuberculosis_H37Rv DLELIGRIGWTCRDVWWAATQDPRAWAALPRAGAVIFATGGMDSLPSVLP
MMAR_3746|M.marinum_M DLELIGRIGWTCRDVWWAATQDPRSWAALPRAGAVIFATSGMDSLPSVLP
MUL_3690|M.ulcerans_Agy99 DLELIGRIGWTCRDVWWAATQDPRSWAALPRAGAVVFATSGMDSLPSVLP
MAV_1753|M.avium_104 DVELIGRIGWTCRDVWWAATQDPRAWAALPRAGAVIFATGGMDSLPSVLP
MAB_1624|M.abscessus_ATCC_1997 DLELIARIGWTCRDVWWAAIQDPRAWAAVPTAGAVIFATGGMDSLPSPLP
*:***.*****.******* ****:***:* ****:***.******* **
MSMEG_4578|M.smegmatis_MC2_155 TALREMIRYVRPPWLRRWVRDGYGWVQPRLSPIARSALPPHVTVEYLEMT
TH_0786|M.thermoresistible__bu TALRELIRYVRPPRLRRWVRDGYGWLQPRLSPYARSALPPHVSVQYLEMT
Mflv_2656|M.gilvum_PYR-GCK TALRELIRYVRPPRLRRWARDGYGWLQPRLSPIARPALPPHLTVEYLEMT
Mvan_3923|M.vanbaalenii_PYR-1 TALRELIRYVRPPRVRRWVRDGYGWVQPKLSPVSRSALPPQLTAEYLEMT
Mb2441c|M.bovis_AF2122/97 TALRELIRYVRPSWLRRWVRDGYAWVQPRLSPVARAALPPHLTAEYLEKT
Rv2418c|M.tuberculosis_H37Rv TALRELIRYVRPSWLRRWVRDGYAWVQPRLSPVARAALPPHLTAEYLEKT
MMAR_3746|M.marinum_M TALRELIRYVRPPWLRRWVRDGYAWVQPRLSPVARSALPADLSAEYLEQT
MUL_3690|M.ulcerans_Agy99 TALRELIRYVRPPWLRRWVRDGYAWVQPRLSPVARSALPADLSAEYLEQT
MAV_1753|M.avium_104 TALRELIRYVRPPRLRRWVRDGYGWLQPRLSPVARSALPPHLSVDYLEQT
MAB_1624|M.abscessus_ATCC_1997 TALRESIRLIRPAKLRSWVREGYGWLQPRLSPIARVALPPKLTVEYLEKT
***** ** :**. :* *.*:**.*:**:*** :* ***..::.:*** *
MSMEG_4578|M.smegmatis_MC2_155 RNAIDFNRPGIPVVASLPSVHIAETYGRAHHGREPTVRAITAWAEEHHVP
TH_0786|M.thermoresistible__bu RGAIDFNRPGIPVVAALPSVHIADTYGRAHHGREPTARAITRWADEHDVA
Mflv_2656|M.gilvum_PYR-GCK RAAIDFNRPGIPVVASLPSVHIAPTYGMSHRGRPGTVAAISAWAAEHGVP
Mvan_3923|M.vanbaalenii_PYR-1 RAAIDFNRPGIPVVAALPSVHIAPTYGMAHHGRAGTVAAITEWAAEREVP
Mb2441c|M.bovis_AF2122/97 RGAIDFNRPGIPIIASLPSVHIAETYGKAHHGRAGTVAAITEWAQHHDIP
Rv2418c|M.tuberculosis_H37Rv RGAIDFNRPGIPIIASLPSVHIAETYGKAHHGRAGTVAAITEWAQHHDIP
MMAR_3746|M.marinum_M RSAIDFNRPGIPVVASLPSVHIAETYGKAHHGRPGTVSAITRWAQRHDVP
MUL_3690|M.ulcerans_Agy99 RSAIDFNRPGIPVVASLPSVHIAETYGKAHHGRPGTVSAITRWAQRHDVP
MAV_1753|M.avium_104 RGAIDFNRPGIPIVASLPSVHIADTYGRAHHGRAATAAAITEWAQSHDIP
MAB_1624|M.abscessus_ATCC_1997 RGALDFNRPGLPMVASLPSVHIAESYGRVHSGREATASAIAAWASEHKIP
* *:******:*::*:******* :** * ** *. **: ** : :.
MSMEG_4578|M.smegmatis_MC2_155 LVDLKAAVADEVFGGRGNPDGIHWSFEAHRAVAELMLKGLAEAG---VTQ
TH_0786|M.thermoresistible__bu LVDFKAAVEDHIMSGRGNPDGIHWNFEAHRAVADLMLKALAEAG---VPP
Mflv_2656|M.gilvum_PYR-GCK LVDLKAAVGEEVMSGRGNPDGIHWNFEAHQAVADLMLKGLAEAG---VPA
Mvan_3923|M.vanbaalenii_PYR-1 LVDLKAAVGEEVMSGRGNPDGIHWNFEAHRAVAELMIKGLSEAG---VEV
Mb2441c|M.bovis_AF2122/97 LVDLKAAVAEQILSGYGNRDGIHWNFEAHQAVAELMLKALAEAG---VPN
Rv2418c|M.tuberculosis_H37Rv LVDLKAAVAEQILSGYGNRDGIHWNFEAHQAVAELMLKALAEAG---VPN
MMAR_3746|M.marinum_M LVDLKAAVAEEIMSGRGNPDGIHWNFEAHQGVAELMLKALAEAG---VPS
MUL_3690|M.ulcerans_Agy99 LVDLKAAVAEEIMSGRGNPDGIHWNFEAHQGVAELMLKALAEAG---VPS
MAV_1753|M.avium_104 LVDLKAAVAEHIMSGRGNPDGIHWNFEAHQAVAELMLKALAEALPNTVPP
MAB_1624|M.abscessus_ATCC_1997 VVDLKQGVGEEVFSGRANPDGIHWNFVAHERVAELMIDALQSVL----PE
:**:* .* :.::.* .* *****.* **. **:**:..* ..
MSMEG_4578|M.smegmatis_MC2_155 RDSAT-
TH_0786|M.thermoresistible__bu DGLER-
Mflv_2656|M.gilvum_PYR-GCK SAADGQ
Mvan_3923|M.vanbaalenii_PYR-1 PASDG-
Mb2441c|M.bovis_AF2122/97 EKSRG-
Rv2418c|M.tuberculosis_H37Rv EKSRG-
MMAR_3746|M.marinum_M EKPRS-
MUL_3690|M.ulcerans_Agy99 EKPRS-
MAV_1753|M.avium_104 TEKR--
MAB_1624|M.abscessus_ATCC_1997 LKAR--