For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSETPLIDAMLAEHAELEKQLADPALHADAAAARKAGRRFAMLSPIVATHRKLATARDDLATARELSADD PSFADEVTELESSIAELETQLSDMLAPRDPHDGDDILLEVKSGEGGEESALFAADLARMYIRYAERHGWK VTVLDETESDLGGYKDATLAIASKGDSADGVWSRLKFEGGVHRVQRVPVTESQGRVHTSAAGVLVYPEPE EVEEIQIDESDLRIDVYRSSGKGGQGVNTTDSAVRITHLPTGIVVTCQNERSQLQNKARAMQVLAARLQA LAEEQAQADASAGRASQIRTVDRSERIRTYNFPENRITDHRVGFKAHNLDQVLDGDLDALFDALAAADRK ARLQEA
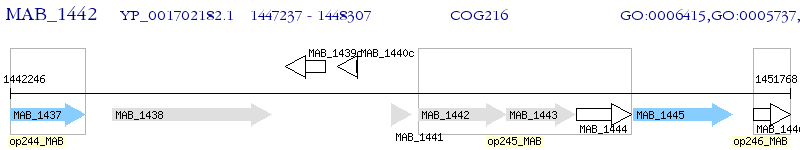
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1442 | prfA | - | 100% (356) | peptide chain release factor 1 |
| M. abscessus ATCC 19977 | MAB_3478c | prfB | 2e-52 | 39.07% (343) | peptide chain release factor 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1331 | prfA | 1e-169 | 84.29% (350) | peptide chain release factor 1 |
| M. gilvum PYR-GCK | Mflv_2307 | prfA | 1e-169 | 85.14% (350) | peptide chain release factor 1 |
| M. tuberculosis H37Rv | Rv1299 | prfA | 1e-169 | 84.29% (350) | peptide chain release factor 1 |
| M. leprae Br4923 | MLBr_01134 | prfA | 1e-169 | 84.42% (353) | peptide chain release factor 1 |
| M. marinum M | MMAR_4098 | prfA | 1e-170 | 85.39% (349) | peptide chain release factor 1 PrfA |
| M. avium 104 | MAV_1516 | prfA | 1e-170 | 84.57% (350) | peptide chain release factor 1 |
| M. smegmatis MC2 155 | MSMEG_4950 | prfA | 1e-174 | 86.36% (352) | peptide chain release factor 1 |
| M. thermoresistible (build 8) | TH_1567 | prfA | 1e-168 | 83.67% (349) | peptide chain release factor 1 |
| M. ulcerans Agy99 | MUL_3965 | prfA | 1e-168 | 83.71% (350) | peptide chain release factor 1 |
| M. vanbaalenii PYR-1 | Mvan_4339 | prfA | 1e-168 | 83.38% (355) | peptide chain release factor 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1331|M.bovis_AF2122/97 --MTQPVQT-IDVLLAEHAELELALADPALHSNPAEARRVGRRFARLAPI
Rv1299|M.tuberculosis_H37Rv --MTQPVQT-IDVLLAEHAELELALADPALHSNPAEARRVGRRFARLAPI
MLBr_01134|M.leprae_Br4923 --MAQPVQS-IDVLLIEHAELELALADPELHSNPAEARKAGRRFARLAPI
MMAR_4098|M.marinum_M --MAAPVQT-IDVLLAEHADLERALADPELHSRPDEARKAGRRFARLAPI
MUL_3965|M.ulcerans_Agy99 --MAAPVQT-IDVLLAEHADLERALADPELHSRPDEARKAGRRFARLARI
MAV_1516|M.avium_104 --MTQPVQT-IDVLLAEHADLERRLSDPDLHSNPDEARKAGRRFARLAPI
MAB_1442|M.abscessus_ATCC_1997 ---MSETPL-IDAMLAEHAELEKQLADPALHADAAAARKAGRRFAMLSPI
Mflv_2307|M.gilvum_PYR-GCK --MAEAAPA-IEAILAEYGELEQRLADPDLHADAGAARKVGRRFAQISPI
Mvan_4339|M.vanbaalenii_PYR-1 --MTETAPA-IEAILAEYGELEQRLADPELHADPAAAKKVGRRFAQISPI
MSMEG_4950|M.smegmatis_MC2_155 MSGTDTAPA-IEALLAEHADLERQLADPALHADAGAARKVGRRFAQVSPI
TH_1567|M.thermoresistible__bu --MTQSAPARIEAVLAEHGDLERQLADPELHADAAKARRAGRRFAQLAPI
. *:.:* *:.:** *:** **: . *::.***** :: *
Mb1331|M.bovis_AF2122/97 VATHRKLTSARDDLETARELVASDESFAAEVAALEARVGELDAQLTDMLA
Rv1299|M.tuberculosis_H37Rv VATHRKLTSARDDLETARELVASDESFAAEVAALEARVGELDAQLTDMLA
MLBr_01134|M.leprae_Br4923 VATHRKLISARDDLQTARELAAGDESFADEIAELESRIAELTTQLTDMLA
MMAR_4098|M.marinum_M VATYRKLVAAREDLETAHELAATDESFAAEAAELEARVAELDSQLTDMLA
MUL_3965|M.ulcerans_Agy99 VATYRKLVAAREDLETAHELAATDESFVAEAAELEARVAELDSQLTDMLA
MAV_1516|M.avium_104 VGTYRKLMAAREDLETARELAADDDSFTAEVADLESRVAQLDTQLTDMLA
MAB_1442|M.abscessus_ATCC_1997 VATHRKLATARDDLATARELSADDPSFADEVTELESSIAELETQLSDMLA
Mflv_2307|M.gilvum_PYR-GCK VATYRKLEAARGDLEAARELAADDASFAAEVDELTDQIGDLDRALTDLLA
Mvan_4339|M.vanbaalenii_PYR-1 VSTYHKLEAARGDLEAARELAADDASFAAEVDELTTKVADLDDRLTDLLA
MSMEG_4950|M.smegmatis_MC2_155 VACYRKLEAARGDLEAARELAADDASFADEVEELTAKVAELDNQLTDLLA
TH_1567|M.thermoresistible__bu VATYRKLEAARGDLEAARELAADDSSFAEEVRELTAKVDELESQLTDLLA
*. ::** :** ** :*:** * * **. * * : :* *:*:**
Mb1331|M.bovis_AF2122/97 PRDPHDADDIVLEVKSGEGGEESALFAADLARMYIRYAERHGWAVTVLDE
Rv1299|M.tuberculosis_H37Rv PRDPHDADDIVLEVKSGEGGEESALFAADLARMYIRYAERHGWAVTVLDE
MLBr_01134|M.leprae_Br4923 PHDPHGPDDIVLEVKSGEGGEESALFAADLARMYIRYAERHGWTVTVLDE
MMAR_4098|M.marinum_M PRDPHDADDIVLEVKSGEGGEESALFAADLARMYIRYAERHGWTVTVLGE
MUL_3965|M.ulcerans_Agy99 PRDPHDADDIVLEVKSGEGGEESALFAADLARMYIRYAERHGWTVTVLGE
MAV_1516|M.avium_104 PRDSHDADDIVLEVKSGEGGEESALFAADLARMYIRYAERHGWTVTVLDE
MAB_1442|M.abscessus_ATCC_1997 PRDPHDGDDILLEVKSGEGGEESALFAADLARMYIRYAERHGWKVTVLDE
Mflv_2307|M.gilvum_PYR-GCK PRDPHDADDIVLEVKSGEGGEESALFAADLARMYIRYAERHGWTVTMLGE
Mvan_4339|M.vanbaalenii_PYR-1 PRDPHDADDIVLEVKSGEGGEESALFAADLARMYIRYAERHGWTVTMLGE
MSMEG_4950|M.smegmatis_MC2_155 PRDPHDADDIVLEVKSGEGGEESALFAADLARMYIRYAERHGWTVTLLDE
TH_1567|M.thermoresistible__bu PRDPHDADDVVLEVKSGEGGEESALFAADLARMYMRYAERRGWSVTILDE
*:*.*. **::***********************:*****:** **:*.*
Mb1331|M.bovis_AF2122/97 TTSDLGGYKDATLAIASK----ADTPDGVWSRMKFEGGVHRVQRVPVTES
Rv1299|M.tuberculosis_H37Rv TTSDLGGYKDATLAIASK----ADTPDGVWSRMKFEGGVHRVQRVPVTES
MLBr_01134|M.leprae_Br4923 TTSDLGGYKDATLAISHKGASDGDAVDGVWSRMKFEGGVHRVQRVPVTES
MMAR_4098|M.marinum_M TTSDLGGYKDATLAIASK----GDTADGVWSRMKFEGGVHRVQRVPVTES
MUL_3965|M.ulcerans_Agy99 TTSDLGGYRDATLAIASK----GDTADGVWSRMKFEGGVHRVQRVPVTES
MAV_1516|M.avium_104 ITSDLGGYKDATLSIRSK----GDSADGVWSRMKFEGGVHRVQRVPVTES
MAB_1442|M.abscessus_ATCC_1997 TESDLGGYKDATLAIASK----GDSADGVWSRLKFEGGVHRVQRVPVTES
Mflv_2307|M.gilvum_PYR-GCK TTSDLGGYKDATLSIASK----GDSADGVWARLKFEGGVHRVQRVPVTES
Mvan_4339|M.vanbaalenii_PYR-1 TTSDLGGYKDATLSIAGK----GDSADGVWSRLKFEGGVHRVQRVPVTES
MSMEG_4950|M.smegmatis_MC2_155 TTSDLGGYKDATLSIRSR----GDSADGVWARLKFEGGVHRVQRVPVTES
TH_1567|M.thermoresistible__bu THSDLGGYKDVTLSIASR----GDSADGVWSRLKFEGGVHRVQRVPVTES
******:*.**:* : .*: ****:*:*****************
Mb1331|M.bovis_AF2122/97 QGRVHTSAAGVLVYPEPEEVGQVQIDESDLRIDVFRSSGKGGQGVNTTDS
Rv1299|M.tuberculosis_H37Rv QGRVHTSAAGVLVYPEPEEVGQVQIDESDLRIDVFRSSGKGGQGVNTTDS
MLBr_01134|M.leprae_Br4923 QGRVHTSAAGVLVYPEPEEIGEVHIDESDLRIDVYRSSGKGGQGVNTTDS
MMAR_4098|M.marinum_M QGRVHTSAAGVLVYPEPEEVGEVQIDESDLRIDVYRSSGKGGQGVNTTDS
MUL_3965|M.ulcerans_Agy99 QGRVHTSAAGVLVYPEPEEVGEVQIDESDLRIDVYRSSGKGGQGVNTTDS
MAV_1516|M.avium_104 QGRVHTSAAGVLVYPEPEEVAEVQIDESDLRIDVFRSSGKGGQGVNTTDS
MAB_1442|M.abscessus_ATCC_1997 QGRVHTSAAGVLVYPEPEEVEEIQIDESDLRIDVYRSSGKGGQGVNTTDS
Mflv_2307|M.gilvum_PYR-GCK QGRVHTSAAGVLVYPEPDEVEQVQIDESDLRVDVYRSSGKGGQGVNTTDS
Mvan_4339|M.vanbaalenii_PYR-1 QGRVHTSAAGVLVYPEPDEVEQVQIDESDLRIDVYRSSGKGGQGVNTTDS
MSMEG_4950|M.smegmatis_MC2_155 QGRVHTSAAGVLVYPEPEEVEEVQIDESDLRIDVYRSSGKGGQGVNTTDS
TH_1567|M.thermoresistible__bu QGRVHTSAAGVLVYPEPEEVEEVQIAESDLRIDVFRSSGKGGQGVNTTDS
*****************:*: :::* *****:**:***************
Mb1331|M.bovis_AF2122/97 AVRITHLPTGIVVTCQNERSQLQNKTRALQVLAARLQAMAEEQALADASA
Rv1299|M.tuberculosis_H37Rv AVRITHLPTGIVVTCQNERSQLQNKTRALQVLAARLQAMAEEQALADASA
MLBr_01134|M.leprae_Br4923 AVRITHLPTGVVVTCQNERSQLQNKTRALQVLAARLQAMAEEQALANASA
MMAR_4098|M.marinum_M AVRITHLPTGIVVTCQNERSQLQNKIRAMQVLGARLQAIAEEQAMADASA
MUL_3965|M.ulcerans_Agy99 AVRITHLPTGIVVTCQNERSQLQNKIRAMQVLGARLQAIAEERAMADASA
MAV_1516|M.avium_104 AVRITHLPTGVVVTCQNERSQLQNKARALQVLAARLQAMAEEQASAEASA
MAB_1442|M.abscessus_ATCC_1997 AVRITHLPTGIVVTCQNERSQLQNKARAMQVLAARLQALAEEQAQADASA
Mflv_2307|M.gilvum_PYR-GCK AVRITHLPTGIVVTCQNERSQLQNKARAMQVLAARLQALAEEQASADASA
Mvan_4339|M.vanbaalenii_PYR-1 AVRITHLPTGIVVTCQNERSQLQNKARAMQVLAARLQALAEEQASADASA
MSMEG_4950|M.smegmatis_MC2_155 AVRITHLPTGIVVTCQNERSQLQNKARAMQVLAARLQALAEEQAEADASA
TH_1567|M.thermoresistible__bu AVRITHLPTGIVVTCQNERSQLQNKIRAMQVLAARLKALAEEQALAEASA
**********:************** **:***.***:*:***:* *:***
Mb1331|M.bovis_AF2122/97 DRASQIRTVDRSERIRTYNFPENRITDHRIGYKSHNLDQVLDGDLDALFD
Rv1299|M.tuberculosis_H37Rv DRASQIRTVDRSERIRTYNFPENRITDHRIGYKSHNLDQVLDGDLDALFD
MLBr_01134|M.leprae_Br4923 DRASQIRTVDRSERIRTYNFPENRITDHRIGYKAHNLDQVLDGDLDALFD
MMAR_4098|M.marinum_M DRASQIRTVDRSERIRTYNFPENRITDHRIGYKSHNLDQVLDGDLDALFD
MUL_3965|M.ulcerans_Agy99 DRASQIRTVDRSERIRTYNFPENRVTDHRIGYKSHNLDQVLDGDLDALFD
MAV_1516|M.avium_104 DRASQIRTVDRSERIRTYNFPENRITDHRIGFKAHNLDQVLDGDLDALFD
MAB_1442|M.abscessus_ATCC_1997 GRASQIRTVDRSERIRTYNFPENRITDHRVGFKAHNLDQVLDGDLDALFD
Mflv_2307|M.gilvum_PYR-GCK DRASQIRTVDRSERIRTYNFPENRIADHRINFKAHNLDQVLDGDLDPLLD
Mvan_4339|M.vanbaalenii_PYR-1 DRASQIRTVDRSERIRTYNYPENRIADHRINYKSHNLDQVLDGDLDPLFD
MSMEG_4950|M.smegmatis_MC2_155 DRASQIRTVDRSERIRTYNFPENRIADHRINFKAHNLDQVLDGDMDALLD
TH_1567|M.thermoresistible__bu DRASQIRTVDRSERIRTYNFPENRITDHRIGYKAHNLDQVLDGDLEPLVE
.******************:****::***:.:*:**********::.*.:
Mb1331|M.bovis_AF2122/97 ALSAADKQSRLRQS-
Rv1299|M.tuberculosis_H37Rv ALSAADKQSRLRQS-
MLBr_01134|M.leprae_Br4923 ALSAADKQSRLQQV-
MMAR_4098|M.marinum_M ALAAADKQARLQQP-
MUL_3965|M.ulcerans_Agy99 ALAAADKQARLQQS-
MAV_1516|M.avium_104 ALAAADKQSRLQQA-
MAB_1442|M.abscessus_ATCC_1997 ALAAADRKARLQEA-
Mflv_2307|M.gilvum_PYR-GCK ALAAADRQARLQNT-
Mvan_4339|M.vanbaalenii_PYR-1 ALAAADKQARLQNT-
MSMEG_4950|M.smegmatis_MC2_155 ALAAADKQSRLQQA-
TH_1567|M.thermoresistible__bu ALIAADRQSRLQQTT
** ***:::**::