For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGAVALGLATIAADGSVLDTWFPAPELTDTPGTPGTERVFEPDAPAELSGLAGSDPDRGVEKVLVRTTIG SLEDKPVDAYDAYLRLHLLSHRLIQPHGASVDGLFGVLTNVVWTNFGPCAVEGFEAVRARLKARAGAHSS VTVFGIDKFPRMVDYVVPTGVRIADADRVRLGAHLAPGTTVMHEGFVNFNAGTLGASMVEGRISAGVVVG DGSDIGGGASIMGTLSGGGSQVISVGQRSLLGANSGLGISLGDDCIIEAGLYVTAGTKVATPDGKTIKAL ELSGANNLLFRRNSQTGAVEVVSRDGGAFELNAALHAN*
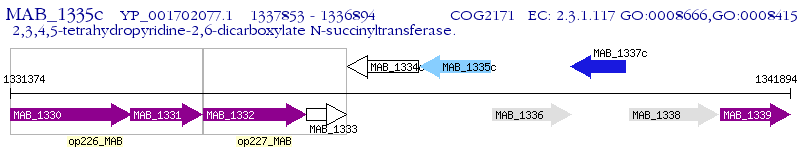
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1335c | - | - | 100% (319) | transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1233c | - | 1e-136 | 74.61% (319) | transferase |
| M. gilvum PYR-GCK | Mflv_2179 | - | 1e-135 | 75.47% (318) | putative transferase |
| M. tuberculosis H37Rv | Rv1201c | - | 1e-136 | 74.61% (319) | transferase |
| M. leprae Br4923 | MLBr_01058 | - | 1e-134 | 72.96% (318) | putative tranferase |
| M. marinum M | MMAR_4236 | - | 1e-140 | 76.80% (319) | transferase |
| M. avium 104 | MAV_1345 | - | 1e-140 | 77.36% (318) | tetrahydropicolinate succinylase |
| M. smegmatis MC2 155 | MSMEG_5104 | - | 1e-132 | 73.67% (319) | tetrahydropicolinate succinylase |
| M. thermoresistible (build 8) | TH_1841 | - | 1e-141 | 78.26% (322) | PROBABLE TRANSFERASE |
| M. ulcerans Agy99 | MUL_0953 | - | 1e-140 | 76.49% (319) | transferase |
| M. vanbaalenii PYR-1 | Mvan_4519 | - | 1e-136 | 76.47% (323) | putative transferase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01058|M.leprae_Br4923 MTGTAGGVGIGLATLAADGSILDTWFPAPKLTES----GTSVTSQLAMSD
MAV_1345|M.avium_104 MTTAAGAAGVGLATLAADGSILDTWFPAPELTGP----GTSGTSRLALSD
Mb1233c|M.bovis_AF2122/97 MSTVTGAAGIGLATLAADGSVLDTWFPAPELTES----GTSATSRLAVSD
Rv1201c|M.tuberculosis_H37Rv MSTVTGAAGIGLATLAADGSVLDTWFPAPELTES----GTSATSRLAVSD
MMAR_4236|M.marinum_M ---MTGAAGIGLATLASDGSVLDTWFPAPELTES----GTSATSRLALSD
MUL_0953|M.ulcerans_Agy99 ---MTGAAGIGLATLASDGSILDTWFPAPELTES----GTSATSRLALSD
Mflv_2179|M.gilvum_PYR-GCK MTSATGASGVGIATIAADGSVLDTWFPAPELTAD----GPSGTSRLSVAE
Mvan_4519|M.vanbaalenii_PYR-1 MTSAS---GIGVATIAADGSVLDTWFPAPELTDG----GEAGTVRLSVAE
MSMEG_5104|M.smegmatis_MC2_155 MTAAS---GIGLATITADGTVLDTWFPAPELSAS----GPAGTARLTGDD
TH_1841|M.thermoresistible__bu VTSAA---GIGLATIAADGTVLDTWFPAPELGAATAEGGAAGTTRLSGAD
MAB_1335c|M.abscessus_ATCC_199 MSGAV---ALGLATIAADGSVLDTWFPAPELTDTP---GTPGTERVFEPD
.:*:**:::**::********:* * . * :: :
MLBr_01058|M.leprae_Br4923 VPYELAVLTGRDDDRGTETIAVRTVIGSLDEVAADAYDAYLRLHMLSHRL
MAV_1345|M.avium_104 VPPELTALIGRDDDRGTETVAVRTVIGSLDDVAADAYDAYLRLHLLSHRL
Mb1233c|M.bovis_AF2122/97 VPVELAALIGRDDDRRTETIAVRTVIGSLDDVAADPYDAYLRLHLLSHRL
Rv1201c|M.tuberculosis_H37Rv VPVELAALIGRDDDRRTETIAVRTVIGSLDDVAADPYDAYLRLHLLSHRL
MMAR_4236|M.marinum_M VPAELAALVGRDDDRGTEVVAVRTVIGSLDDVAADAYDAYLRLHLLSHRL
MUL_0953|M.ulcerans_Agy99 VPAELAALVGGDDDRGTEVVAVRTVIGSLDDVAADAYDAYLRLHLLSHRL
Mflv_2179|M.gilvum_PYR-GCK LPPELAALTGRDEDRDVDVVAVRTVIASLADKAVDAYDAYLRLHLLSHRL
Mvan_4519|M.vanbaalenii_PYR-1 VPAELAALAGRDEDRDVDVVVVRTVIASLDDKAVDAYDAYLRLHLLSHRL
MSMEG_5104|M.smegmatis_MC2_155 VPADLAALTGKDEDRDVEVVAVRTTIADLNDKPADTHDVWLRLHLLSHRL
TH_1841|M.thermoresistible__bu APAELAALTGRDDDRGVETVVVRTMIASLDDKPADAHDAYLRLHLLSHRL
MAB_1335c|M.abscessus_ATCC_199 APAELSGLAGSDPDRGVEKVLVRTTIGSLEDKPVDAYDAYLRLHLLSHRL
* :*: * * * ** .: : *** *..* : ..*.:*.:****:*****
MLBr_01058|M.leprae_Br4923 VAPHGLNADGLFGVLTNVVWTSRGPCAIDGFDTVRAQLRRHG----PVTI
MAV_1345|M.avium_104 VAPHGLNAGGLFGVLTNVVWTNRGPCAIEGFEAVRARLRRHG----PVTV
Mb1233c|M.bovis_AF2122/97 VAPHGLNAGGLFGVLTNVVWTNHGPCAIDGFEAVRARLRRRG----PVTV
Rv1201c|M.tuberculosis_H37Rv VAPHGLNAGGLFGVLTNVVWTNHGPCAIDGFEAVRARLRRRG----PVTV
MMAR_4236|M.marinum_M VAPHGLNAGGLFGVLTNVVWTNRGPCAIEGFEAVRARLRRHG----AVTV
MUL_0953|M.ulcerans_Agy99 VAPHGLNAGGLFGVLTNVVWTNRGPCAIEGFEAVRARLRRHG----AVTV
Mflv_2179|M.gilvum_PYR-GCK VAPHGLNADGLFGVLTNVVWTSHGPCSVDDFERVRARLRRRG----QVAV
Mvan_4519|M.vanbaalenii_PYR-1 VAPHGLNADGLFGVLTNVVWTSHGPCAVDGFETVRARLRRRG----HVAV
MSMEG_5104|M.smegmatis_MC2_155 TKPHEANLDGIFGLLSNVVWTNFGPCAVEGFETTRARLRKRG----AVAV
TH_1841|M.thermoresistible__bu IKPHGANMDGIFGVLANVVWTNHGPCAVEGFETVRARLRRRG----PVTV
MAB_1335c|M.abscessus_ATCC_199 IQPHGASVDGLFGVLTNVVWTNFGPCAVEGFEAVRARLKARAGAHSSVTV
** . .*:**:*:*****. ***:::.*: .**:*: :. *::
MLBr_01058|M.leprae_Br4923 YGVDKFPRMVDYVVPTGVRIANADRVRLGAHLAPGTTVMHEGFVNYNAGT
MAV_1345|M.avium_104 YGVDKFPRMVDYVLPTGVRIADADRVRLGAHLAPGTTVMHEGFVNYNAGT
Mb1233c|M.bovis_AF2122/97 YGVDKFPRMVDYVVPTGVRIADADRVRLGAHLAPGTTVMHEGFVNYNAGT
Rv1201c|M.tuberculosis_H37Rv YGVDKFPRMVDYVVPTGVRIADADRVRLGAHLAPGTTVMHEGFVNYNAGT
MMAR_4236|M.marinum_M YGVDKFPRMVDYVMPSGVRIADADRVRLGAHLAPGTTVMHEGFVNYNAGT
MUL_0953|M.ulcerans_Agy99 YGVDKFPRMVDYVMPSGVRIADADRVRLGAHLAPGTTVMHEGFVNYNAGT
Mflv_2179|M.gilvum_PYR-GCK YGVDKFPRMVDYVLPSGVRIADADRVRLGAHLAPGTTVMHEGFVNFNAGT
Mvan_4519|M.vanbaalenii_PYR-1 YGVDKFPRMVDYVLPSGVRIADADRVRLGAHLAPGTTVMHEGFVNFNAGT
MSMEG_5104|M.smegmatis_MC2_155 YGIDKFPRMVDYVTPSGVRIADADRVRLGAHLASGTTVMHEGFVNFNAGT
TH_1841|M.thermoresistible__bu YGVDKFPRMVDYVTPTGVRIADADRVRLGAHLAPGTTVMHEGFVNFNAGT
MAB_1335c|M.abscessus_ATCC_199 FGIDKFPRMVDYVVPTGVRIADADRVRLGAHLAPGTTVMHEGFVNFNAGT
:*:********** *:*****:***********.***********:****
MLBr_01058|M.leprae_Br4923 LGASMVEGRISAGVVVGEGSHVGGGASIMGTLSGGGTQVISIGKRCLLGA
MAV_1345|M.avium_104 LGASMVEGRISAGVVVGDGSDIGGGASIMGTLSGGGTEVISIGKRCLLGA
Mb1233c|M.bovis_AF2122/97 LGASMVEGRISAGVVVGDGSDVGGGASIMGTLSGGGTHVISIGKRCLLGA
Rv1201c|M.tuberculosis_H37Rv LGASMVEGRISAGVVVGDGSDVGGGASIMGTLSGGGTHVISIGKRCLLGA
MMAR_4236|M.marinum_M LGASMVEGRISAGVVVGDGSDIGGGASIMGTLSGGGTQIISMGKRCLLGA
MUL_0953|M.ulcerans_Agy99 LGASMVEGRISAGVVVGDGSDIGGGASIMGTLSGGGTQIISMGNRCLLGA
Mflv_2179|M.gilvum_PYR-GCK LGSSMVEGRISAGVVVDDGSDVGGGASIMGTLSGGGTEVISVGKRCLLGA
Mvan_4519|M.vanbaalenii_PYR-1 LGSSMVEGRISAGVVVDDGSDVGGGASIMGTLSGGGTQVISVGKRCLLGA
MSMEG_5104|M.smegmatis_MC2_155 LGTSMVEGRISAGVVVDDGSDIGGGASIMGTLSGGGKEVIKVGKRCLLGA
TH_1841|M.thermoresistible__bu LGTSMVEGRISAGVVVDDGSDLGGGASIMGTLSGGGKEVISVGKRCLLGA
MAB_1335c|M.abscessus_ATCC_199 LGASMVEGRISAGVVVGDGSDIGGGASIMGTLSGGGSQVISVGQRSLLGA
**:*************.:**.:**************..:*.:*:*.****
MLBr_01058|M.leprae_Br4923 NSGLGISLGDDCIVEAGLYVTTGTKVNTPEGK----SVKARALSGGSNLL
MAV_1345|M.avium_104 NAGLGISLGDDCVVEAGLYVTAGTKVITPEGK----QLKARELSGGNNLL
Mb1233c|M.bovis_AF2122/97 NSGLGISLGDDCVVEAGLYVTAGTRVTMPDSN----SVKARELSGSSNLL
Rv1201c|M.tuberculosis_H37Rv NSGLGISLGDDCVVEAGLYVTAGTRVTMPDSN----SVKARELSGSSNLL
MMAR_4236|M.marinum_M NAGLGISLGDDCVVEAGLYVTAGTKVTTPDGI----TVRARELSGSSNLL
MUL_0953|M.ulcerans_Agy99 NAGLGISLGDDCVVEAGLYVTAGTKVTTPDGI----TVRARELSGSSNLL
Mflv_2179|M.gilvum_PYR-GCK NAGLGISLGDDCVIEAGLYVTAGTKVTTSDGQ----TVKARDLSGASNLL
Mvan_4519|M.vanbaalenii_PYR-1 NAGLGISLGDDCVIEAGLYVTAGTKVTLLDGDRTATTVKARELSGANNLL
MSMEG_5104|M.smegmatis_MC2_155 NSGLGISLGDDCVVEAGLYVTGGTKVTTADGQ----VTKAIELSGASNLL
TH_1841|M.thermoresistible__bu NSGLGISLGDDCVVEAGLYVTAGTKVTTADGK----TVKARELSGADNLL
MAB_1335c|M.abscessus_ATCC_199 NSGLGISLGDDCIIEAGLYVTAGTKVATPDGK----TIKALELSGANNLL
*:**********::******* **:* :. :* ***..***
MLBr_01058|M.leprae_Br4923 FRRNSVTGAVEVVARDGRGSTLNEALHTN
MAV_1345|M.avium_104 FRRNSLTGAVEVVARGGTGITLNEDLHAN
Mb1233c|M.bovis_AF2122/97 FRRNSVSGAVEVLARDGQGIALNEDLHAN
Rv1201c|M.tuberculosis_H37Rv FRRNSVSGAVEVLARDGQGIALNEDLHAN
MMAR_4236|M.marinum_M FRRNSVTGAVEVVARDGQGIALNEDLHAN
MUL_0953|M.ulcerans_Agy99 FRRNSVTGAVEVVARDGQGIALNEVLHAN
Mflv_2179|M.gilvum_PYR-GCK FRRNSVTGAVEVVKRDGTGITLNEALHAN
Mvan_4519|M.vanbaalenii_PYR-1 FRRNSVTGAVEVVKRDGTGITLNEALHAN
MSMEG_5104|M.smegmatis_MC2_155 FRRNSLSGAVEVVKRDGTGITLNEALHAN
TH_1841|M.thermoresistible__bu FRRNSVTGAVEVVKRDGTGITLNEALHAN
MAB_1335c|M.abscessus_ATCC_199 FRRNSQTGAVEVVSRDGGAFELNAALHAN
***** :*****: *.* . ** **:*