For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTSASSDIAQGAQYAETASAAYRSALQVIETIEPRIADATRKELADQRDSLKLIASENYASPAVLLTMG TWLSDKYAEGTIGHRFYAGCQNIDTVEALAAEHARELFGAPYAYAQPHSGIDANLVAYWAILATRVEAPA LADKGVRNVNDLSETDWEELRHQYGNQRLMGMSLDAGGHLTHGFRPNISGKMFHQRSYGTDPETGLLDYD ALAAAAREFKPLVLVGGYSAYPRRVNFAKLREIADEVGATLFVDMAHFAGLVAGKVFTGDENPVPHAHIT TTTTHKSLRGPRGGLVLATAEYSDAVDKGCPMVLGGPLSHVMAAKAVALAEARQPSFQAYAQRVADNAKS LAEGFLKRGARLVTGGTDNHLVLLDVQSFGLTGRQAESALLDAGVVTNRNAIPADPNGAWYTSGIRFGTP ALTSRGFGADEFDKVAELVVDVLTNTEADGSSKAKYTLADAVAERVKAASAELLAANPLYPGLTL
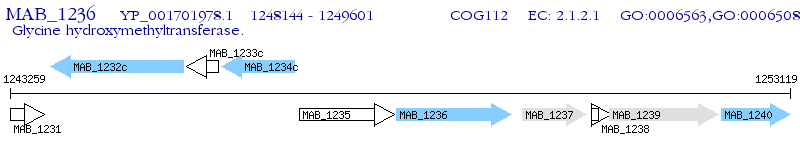
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1236 | - | - | 100% (485) | serine hydroxymethyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1123 | glyA | 1e-97 | 43.98% (457) | serine hydroxymethyltransferase |
| M. gilvum PYR-GCK | Mflv_2060 | glyA | 0.0 | 83.57% (487) | serine hydroxymethyltransferase |
| M. tuberculosis H37Rv | Rv1093 | glyA | 3e-97 | 43.76% (457) | serine hydroxymethyltransferase |
| M. leprae Br4923 | MLBr_01953 | glyA | 2e-94 | 44.74% (418) | serine hydroxymethyltransferase |
| M. marinum M | MMAR_4375 | glyA | 3e-96 | 43.98% (457) | serine hydroxymethyltransferase 1 GlyA1 |
| M. avium 104 | MAV_1215 | glyA | 5e-93 | 43.33% (457) | serine hydroxymethyltransferase |
| M. smegmatis MC2 155 | MSMEG_5249 | glyA | 0.0 | 84.30% (484) | serine hydroxymethyltransferase |
| M. thermoresistible (build 8) | TH_2160 | glyA | 0.0 | 84.30% (484) | serine hydroxymethyltransferase |
| M. ulcerans Agy99 | MUL_0188 | glyA | 7e-96 | 43.98% (457) | serine hydroxymethyltransferase |
| M. vanbaalenii PYR-1 | Mvan_4656 | glyA | 0.0 | 83.95% (486) | serine hydroxymethyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4375|M.marinum_M ------------------------------MEAPLAEVDPDIAALLGKEL
MUL_0188|M.ulcerans_Agy99 ------------------------------MEAPLAEVDPDIAALLGKEL
MAV_1215|M.avium_104 -------------MTAAPDARPTASDTASLMSAPLADIDPDIAGLLGQEL
Mb1123|M.bovis_AF2122/97 ------------------------------MSAPLAEVDPDIAELLAKEL
Rv1093|M.tuberculosis_H37Rv ------------------------------MSAPLAEVDPDIAELLAKEL
MLBr_01953|M.leprae_Br4923 ------------------------------MVAPLAEVDPDIAELLGKEL
Mflv_2060|M.gilvum_PYR-GCK -------MTSVSASGQGAEYAATASEAYQAALRVIETVEPRIAEATRKEL
Mvan_4656|M.vanbaalenii_PYR-1 -------MTSSSPSGQGADYAATASEAYRAALRVIESVEPRVAEATRKEL
MSMEG_5249|M.smegmatis_MC2_155 MAADPSSNSSSVPAANGADYADTASAAYQAALQVIESVEPRVAAATRKEL
TH_2160|M.thermoresistible__bu --MTSDSTTTASAATPGAEYADTASAAYRAALQVIESVEPRIAEATRKEL
MAB_1236|M.abscessus_ATCC_1997 -----MTTSASSDIAQGAQYAETASAAYRSALQVIETIEPRIADATRKEL
: ::* :* :**
MMAR_4375|M.marinum_M GRQRDTLEMIASENFVPRAVLQAQGSVLTNKYAEGLPGRRYYGGCEHVDV
MUL_0188|M.ulcerans_Agy99 GRQRDTLEMIASENFVPRAVLQVQGSVLTNKYAEGLPGRRYYGGCEHVDV
MAV_1215|M.avium_104 GRQRDTLEMIASENFVPRAVLQAQGSVLTNKYAEGLPGRRYYGGCEYVDV
Mb1123|M.bovis_AF2122/97 GRQRDTLEMIASENFAPRAVLQAQGSVLTNKYAEGLPGRRYYGGCEHVDV
Rv1093|M.tuberculosis_H37Rv GRQRDTLEMIASENFVPRAVLQAQGSVLTNKYAEGLPGRRYYGGCEHVDV
MLBr_01953|M.leprae_Br4923 GRQRDTLEMIASENFVPRSVLQAQGSVLTNKYAEGLPGRRYYDGCEHVDV
Mflv_2060|M.gilvum_PYR-GCK ADQRDSLKLIASENYASPAVLLTMGTWFSDKYAEGTVGHRFYAACQNVDT
Mvan_4656|M.vanbaalenii_PYR-1 ADQRGSLKLIASENYASPAVLLTMGTWFSDKYAEGTVGHRFYAACQNVDT
MSMEG_5249|M.smegmatis_MC2_155 ADQRDSLKLIASENYASPAVLLTMGTWFSDKYAEGTIGHRFYAGCQNVDT
TH_2160|M.thermoresistible__bu ADQRESLKLIASENYASPATLLTMGTWLSDKYAEGTVGHRFYAGCQNVDT
MAB_1236|M.abscessus_ATCC_1997 ADQRDSLKLIASENYASPAVLLTMGTWLSDKYAEGTIGHRFYAGCQNIDT
. ** :*::*****:.. :.* . *: :::***** *:*:* .*: :*.
MMAR_4375|M.marinum_M VENIARDRAKALFGADFANVQPHSGAQANAAVLHALMSP-----------
MUL_0188|M.ulcerans_Agy99 VENIARDRAKALFGADFADVQPHSGAQANAAVLHALMSP-----------
MAV_1215|M.avium_104 VENIARDRAKALFGADFANVQPHSGAQANAAVLHALMTP-----------
Mb1123|M.bovis_AF2122/97 VENLARDRAKALFGAEFANVQPHSGAQANAAVLHALMSP-----------
Rv1093|M.tuberculosis_H37Rv VENLARDRAKALFGAEFANVQPHSGAQANAAVLHALMSP-----------
MLBr_01953|M.leprae_Br4923 VENIARDRAKALFGADFANVQPHSGAQANAAVLHALMSP-----------
Mflv_2060|M.gilvum_PYR-GCK VEALAAEHARELFGAPYAYAQPHSGIDANLVAYWAILATRVEAPGLAEMG
Mvan_4656|M.vanbaalenii_PYR-1 VEALAAEHARELFGAPYAYAQPHSGIDANLVAYWAILATRVEAPGLAELG
MSMEG_5249|M.smegmatis_MC2_155 VESVAAEHARELFGAPYAYVQPHSGIDANLVAFWAILATRVEAPELANFG
TH_2160|M.thermoresistible__bu VESVAAEHARELFGAPYAYVQPHSGIDANLVAFWAILATRVEAPALAEHG
MAB_1236|M.abscessus_ATCC_1997 VEALAAEHARELFGAPYAYAQPHSGIDANLVAYWAILATRVEAPALADKG
** :* ::*: **** :* .***** :** .. *:::.
MMAR_4375|M.marinum_M --------------------GERLLGLDLANGGHLTHGMRLNFSGKLYEN
MUL_0188|M.ulcerans_Agy99 --------------------GERLLGLDLANGGHLTHGMRLNFSGKLYEN
MAV_1215|M.avium_104 --------------------GERLLGLDLANGGHLTHGMKLNFSGKLYDV
Mb1123|M.bovis_AF2122/97 --------------------GERLLGLDLANGGHLTHGMRLNFSGKLYEN
Rv1093|M.tuberculosis_H37Rv --------------------GERLLGLDLANGGHLTHGMRLNFSGKLYEN
MLBr_01953|M.leprae_Br4923 --------------------GERLLGLDLANGGHLTHGMRLNFSGKLYET
Mflv_2060|M.gilvum_PYR-GCK AKHINDLSEADWEKLRAKLGNQRLLGMSLDTGGHLTHGFRPNISGKMFHQ
Mvan_4656|M.vanbaalenii_PYR-1 AKHVNDLSEGDWEKLRAKLGNQRLLGMSLDTGGHLTHGFRPNISGKMFHQ
MSMEG_5249|M.smegmatis_MC2_155 AKHINDLSEADWETLRNKLGNQRLLGMSLDAGGHLTHGFRPNISGKMFHQ
TH_2160|M.thermoresistible__bu VKHINDLSEADWEKLRNKLGNQRLLGMSLDAGGHLTHGFRPNISGKMFHQ
MAB_1236|M.abscessus_ATCC_1997 VRNVNDLSETDWEELRHQYGNQRLMGMSLDAGGHLTHGFRPNISGKMFHQ
.:**:*:.* *******:: *:***::.
MMAR_4375|M.marinum_M GFYGVDPTTHLIDMDAVRAKALEFRPKVIIAGWSAYPRVLDFAAFRSIAD
MUL_0188|M.ulcerans_Agy99 GFYGVDPTTHLIDMDAVRAKALEFRPKVIIAGWSAYPRVLDFAAFRSIAD
MAV_1215|M.avium_104 GFYGVDPTTHLIDMDAVRAKALEFRPKVIIAGWSAYPRVLDFAAFASIAD
Mb1123|M.bovis_AF2122/97 GFYGVDPATHLIDMDAVRATALEFRPKVIIAGWSAYPRVLDFAAFRSIAD
Rv1093|M.tuberculosis_H37Rv GFYGVDPATHLIDMDAVRATALEFRPKVIIAGWSAYPRVLDFAAFRSIAD
MLBr_01953|M.leprae_Br4923 GFYGVDATTHLIDMDAVRAKALEFRPKVLIAGWSAYPRILDFAAFRSIAD
Mflv_2060|M.gilvum_PYR-GCK RQYGTDPTTGFIDYDAVAASAREFKPLVLVAGYSAYPRRVNFAKMREIAD
Mvan_4656|M.vanbaalenii_PYR-1 RQYGTDPATGFIDYDAVAAAAREFKPLVLVAGYSAYPRRVNFAKMREIAD
MSMEG_5249|M.smegmatis_MC2_155 RSYGTNPETGFLDYDAVAAAAREFKPLVLVAGYSAYPRRVNFAKMREIAD
TH_2160|M.thermoresistible__bu RSYGTDPQTGLLDYDAVAAAAREFKPLIIVAGYSAYPRRINFAKMREIAD
MAB_1236|M.abscessus_ATCC_1997 RSYGTDPETGLLDYDALAAAAREFKPLVLVGGYSAYPRRVNFAKLREIAD
**.:. * ::* **: * * **:* :::.*:***** ::** : .***
MMAR_4375|M.marinum_M EVGAQLFVDMAHFAGLVAAGLHPS---PVPHADVVSTTIHKTLGGGRSGM
MUL_0188|M.ulcerans_Agy99 EVGAQLFVDMAHFAGLVAAGLHPS---PVPHADVVSTTIHKTLGGGRSGM
MAV_1215|M.avium_104 EVGAKLWVDMAHFAGLVAAGLHPS---PVPHADVVSTTVHKTLGGPRSGL
Mb1123|M.bovis_AF2122/97 EVGAKLLVDMAHFAGLVAAGLHPS---PVPHADVVSTTVHKTLGGGRSGL
Rv1093|M.tuberculosis_H37Rv EVGAKLLVDMAHFAGLVAAGLHPS---PVPHADVVSTTVHKTLGGGRSGL
MLBr_01953|M.leprae_Br4923 EVGAKLWVDMAHFAGLVAVGLHPS---PVPHADVVSTTVHKTLGGGRSGL
Mflv_2060|M.gilvum_PYR-GCK EVGATLMVDMAHFAGLVAGKVFTGDEDPVPHAHVVTTTTHKSLRGPRGGL
Mvan_4656|M.vanbaalenii_PYR-1 EVGATLMVDMAHFAGLVAGKVFTGDEDPVPHAHITTTTTHKSLRGPRGGL
MSMEG_5249|M.smegmatis_MC2_155 EVGATLMVDMAHFAGLVAGKVFTGDEDPVPHAHVTTTTTHKSLRGPRGGM
TH_2160|M.thermoresistible__bu EVGATLMVDMAHFAGLVAGKVFTGDEDPVPHAHVTTTTTHKSLRGPRGGM
MAB_1236|M.abscessus_ATCC_1997 EVGATLFVDMAHFAGLVAGKVFTGDENPVPHAHITTTTTHKSLRGPRGGL
**** * *********** :... *****.:.:** **:* * *.*:
MMAR_4375|M.marinum_M ILGKQEFAKAINSAVFPGQQGGPLMHVIAGKAVALKIAGTPEFADRQRRT
MUL_0188|M.ulcerans_Agy99 ILGKQEFAKAINSAVFPGQQGGPLMHVIAGKAVALKIAGTPEFADRQRRT
MAV_1215|M.avium_104 ILGKQEYAKSINSAVFPGQQGGPLMHVIAAKAVALKIAGTEEFADRQRRT
Mb1123|M.bovis_AF2122/97 IVGKQQYAKAINSAVFPGQQGGPLMHVIAGKAVALKIAATPEFADRQRRT
Rv1093|M.tuberculosis_H37Rv IVGKQQYAKAINSAVFPGQQGGPLMHVIAGKAVALKIAATPEFADRQRRT
MLBr_01953|M.leprae_Br4923 ILGKQEFATAINSAVFPGQQGGPLMHVIAGKAVALKIATTPEFTDRQQRT
Mflv_2060|M.gilvum_PYR-GCK VLATEEFAPAVDKG-CPMVLGGPLSHVMAAKAVALAEARQPEFRTYAQAV
Mvan_4656|M.vanbaalenii_PYR-1 VLATEEYAPAVDKG-CPMVLGGPLSHVMAAKAVALAEARRPEFRAYAQAV
MSMEG_5249|M.smegmatis_MC2_155 VLATEEYAPAVDKG-CPMVLGGPLSHVMAAKAVALAEARQPAFQQYAQQV
TH_2160|M.thermoresistible__bu VLATEEYAPAVDKG-CPMVLGGPLSHVMAAKAVALAEARQPSFRAYAQRV
MAB_1236|M.abscessus_ATCC_1997 VLATAEYSDAVDKG-CPMVLGGPLSHVMAAKAVALAEARQPSFQAYAQRV
::.. ::: :::.. * **** **:*.***** * * : .
MMAR_4375|M.marinum_M LAGARIVADRLMASDVAKAGVSVVSGGTDVHLVLVDLRDSPLDGQAAEDL
MUL_0188|M.ulcerans_Agy99 LAGARIVADRLMASDVAKAGVSVVSGGTDVHLVLVDLRDSPLDGQAAEDL
MAV_1215|M.avium_104 LSGARILAERLSGADVAAAGVSVVSGGTDVHLVLVDLRNSELDGQAAEDL
Mb1123|M.bovis_AF2122/97 LSGARIIADRLMAPDVAKAGVSVVSGGTDVHLVLVDLRDSPLDGQAAEDL
Rv1093|M.tuberculosis_H37Rv LSGARIIADRLMAPDVAKAGVSVVSGGTDVHLVLVDLRDSPLDGQAAEDL
MLBr_01953|M.leprae_Br4923 LAGARILADRLTAADVTKAGVSVVSGGTDVHLVLVDLRNSPFDGQAAEDL
Mflv_2060|M.gilvum_PYR-GCK ADNAQTLAD-----GFVKRDAGLVTGGTDNHLVLLDVTSFGLTGRQAESA
Mvan_4656|M.vanbaalenii_PYR-1 ADNAQALAD-----GFIKRDGSLVTGGTDNHLVLLDVTSFGLTGRQAESA
MSMEG_5249|M.smegmatis_MC2_155 ADNAQALAD-----GFVKRDAGLVTGGTDNHIVLLDVTSFGLTGRQAESA
TH_2160|M.thermoresistible__bu ADNAKAFAD-----GLLKRGARLVTGGTDNHIVLLDVTSFGLTGRQAEAA
MAB_1236|M.abscessus_ATCC_1997 ADNAKSLAE-----GFLKRGARLVTGGTDNHLVLLDVQSFGLTGRQAESA
.*: .*: .. . :*:**** *:**:*: . : *: **
MMAR_4375|M.marinum_M LHEAGITVNRNAVPNDPRPPMVTSGLRIGTPALATRGFGDAEFSEVADVI
MUL_0188|M.ulcerans_Agy99 LHEAGITVNRNAVPNDPRPPMVTSGLRIGTPALATRGFGDAEFSEVADVI
MAV_1215|M.avium_104 LHEIGITVNRNAVPNDPRPPMVTSGLRVGTPALATRGFGDAEFSEVADVI
Mb1123|M.bovis_AF2122/97 LHEVGITVNRNAVPNDPRPPMVTSGLRIGTPALATRGFGDTEFTEVADII
Rv1093|M.tuberculosis_H37Rv LHEVGITVNRNAVPNDPRPPMVTSGLRIGTPALATRGFGDTEFTEVADII
MLBr_01953|M.leprae_Br4923 LHEVGITVNRNVVPNDPRPPMVTSGLRIGTPALATRGFGEAEFTEVADII
Mflv_2060|M.gilvum_PYR-GCK LLDSGIVTNRNAIPADPNGAWYTSGIRFGSPALTTRGFGADEFDRVSELV
Mvan_4656|M.vanbaalenii_PYR-1 LLDSGIVTNRNAVPADPNGAWYTSGIRFGSPALTTRGFGADDFDRVAELV
MSMEG_5249|M.smegmatis_MC2_155 LLDAGIVTNRNSIPADPNGAWYTSGVRLGTPALTSRGFGADDFDRVAELI
TH_2160|M.thermoresistible__bu LLESGVVTNRNAIPADPNGAWYTSGIRFGTPALTSRGFDAADFDRVAELV
MAB_1236|M.abscessus_ATCC_1997 LLDAGVVTNRNAIPADPNGAWYTSGIRFGTPALTSRGFGADEFDKVAELV
* : *:..*** :* **. . ***:*.*:***::***. :* .*::::
MMAR_4375|M.marinum_M AGVLAAGR----------------SADVAALRARVTRLARDFPLYEGLEE
MUL_0188|M.ulcerans_Agy99 AGALAAGR----------------SADVAALRARVTRLARDFPLYEGLEE
MAV_1215|M.avium_104 ATALAGGR----------------GADLAALRDRVTRLARDFPLYEGLED
Mb1123|M.bovis_AF2122/97 ATALATGS----------------SVDVSALKDRATRLARAFPLYDGLEE
Rv1093|M.tuberculosis_H37Rv ATALATGS----------------SVDVSALKDRATRLARAFPLYDGLEE
MLBr_01953|M.leprae_Br4923 ATVLTTGG----------------SVDVAALRQQVTRLARDFPLYGGLED
Mflv_2060|M.gilvum_PYR-GCK VEVLSNTQPAAGPNGPSKAKYTIADGVAERVHAAASELLEANPLYPGLTL
Mvan_4656|M.vanbaalenii_PYR-1 VEVLSNTQPAAGPNGPSKAKYTLADGTAERVRAAAAELLDANPLYPGLTL
MSMEG_5249|M.smegmatis_MC2_155 VEVLANTQ----PEGTSKAKYKLADGTAERVHAASSELLSANPLYPGLTL
TH_2160|M.thermoresistible__bu VEVLENTQ----PDATSKARYTLAEGTAARVQAAADELLTANPLYPGLTL
MAB_1236|M.abscessus_ATCC_1997 VDVLTNTE----ADGSSKAKYTLADAVAERVKAASAELLAANPLYPGLTL
. .* :: .* *** **
MMAR_4375|M.marinum_M WNLVDR
MUL_0188|M.ulcerans_Agy99 WNLVDR
MAV_1215|M.avium_104 WALVGR
Mb1123|M.bovis_AF2122/97 WSLVGR
Rv1093|M.tuberculosis_H37Rv WSLVGR
MLBr_01953|M.leprae_Br4923 WSLAGR
Mflv_2060|M.gilvum_PYR-GCK ------
Mvan_4656|M.vanbaalenii_PYR-1 ------
MSMEG_5249|M.smegmatis_MC2_155 ------
TH_2160|M.thermoresistible__bu ------
MAB_1236|M.abscessus_ATCC_1997 ------