For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QRPEDRYGTKPATPGRGRWIAIALTALVIVAGVAIAAIGYNRLGPGDVKGELSAYKLVDSSTVSVTASVE RKDPSKPAVCILRARSIDGSETGRREVLVPPSAERSVPVTALVKTTRPPVTGDVYGCSLAIPSYLVAS*
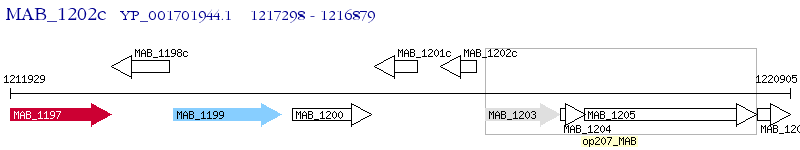
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1202c | - | - | 100% (139) | hypothetical protein MAB_1202c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1110c | - | 2e-36 | 52.94% (136) | hypothetical protein Mb1110c |
| M. gilvum PYR-GCK | Mflv_2049 | - | 2e-43 | 58.39% (137) | hypothetical protein Mflv_2049 |
| M. tuberculosis H37Rv | Rv1081c | - | 2e-36 | 52.94% (136) | hypothetical protein Rv1081c |
| M. leprae Br4923 | MLBr_02392 | - | 5e-35 | 54.14% (133) | hypothetical protein MLBr_02392 |
| M. marinum M | MMAR_4386 | - | 8e-37 | 51.47% (136) | hypothetical protein MMAR_4386 |
| M. avium 104 | MAV_1205 | - | 9e-37 | 50.75% (134) | hypothetical protein MAV_1205 |
| M. smegmatis MC2 155 | MSMEG_5262 | - | 1e-44 | 60.58% (137) | hypothetical protein MSMEG_5262 |
| M. thermoresistible (build 8) | TH_2168 | - | 1e-43 | 55.07% (138) | PROBABLE CONSERVED MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_0199 | - | 8e-37 | 51.47% (136) | hypothetical protein MUL_0199 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1202c|M.abscessus_ATCC_199 -----MQRPEDRYGTKPATPGRGRWIAIALTALVIVAGVAIAAIGYNRLG
MSMEG_5262|M.smegmatis_MC2_155 ----MIERPAARYGSREQPRLSRRWILIAVAALVLVAGVIVAVVAYQRFG
Mflv_2049|M.gilvum_PYR-GCK MQNEMIERPTARYGRQTMSRRTRRWIAAGLTALVVVIGVAVAAVAFSRFG
TH_2168|M.thermoresistible__bu ----MIERPTARYGRQPTSARPRRWAVIGVTVLAVIVGLAVAVIGYRQLG
Mb1110c|M.bovis_AF2122/97 MTHTPIPRPDARYGRPRLSRRARRRVAIALGVLVAAAGIVIAVIGYQRIS
Rv1081c|M.tuberculosis_H37Rv MTHTPIPRPDARYGRPRLSRRARRRVAIALGVLVAAAGIVIAVIGYQRIS
MMAR_4386|M.marinum_M MNPAPISRPEARYGRSRLSGRSRRYILIALAVLVVVAGTVLAVIGYQRIV
MUL_0199|M.ulcerans_Agy99 MNPAPISRPEARYGRSRLSGRSRRYIFIALAVLVVVAGTVLAVIGYQRIV
MLBr_02392|M.leprae_Br4923 MNQAFTPLTETRYGRSRLPGRSRHRGVIALTVLAVAASTGIAVIGYQRLG
MAV_1205|M.avium_104 MTET---RPNERYGRSALSRIPRRWVIAALGVLVVAAGLAVAVVGYHRFG
. *** . : .: .*. . :*.:.: ::
MAB_1202c|M.abscessus_ATCC_199 PGDVKGELSAYKLVDSSTVSVTASVERKDPSKPAVCILRARSIDGSETGR
MSMEG_5262|M.smegmatis_MC2_155 SGEVKGELTGYQLLDDETVQVTIGVERPDPSKPVVCIVRARSKDGSETGR
Mflv_2049|M.gilvum_PYR-GCK SSDVKGEMAGYELVDPHTVDVTISVTRDDPSRPAVCIVRARSRDGDETGR
TH_2168|M.thermoresistible__bu SPDVEGELSGYRVLDDRTVEVTIAVTRKDPARPVVCIVRARSLDGSETGR
Mb1110c|M.bovis_AF2122/97 TSAVTGSLVGYRLVDDETASVTISVTRSDPSRPVACIVRVRATNGSETGR
Rv1081c|M.tuberculosis_H37Rv TSAVTGSLVGYRLVDDETASVTISVTRSDPSRPVACIVRVRATNGSETGR
MMAR_4386|M.marinum_M TNAVTGKLAGYRVIDEQTASVTISVTRSDPSRPVDCIVRVRAKDGSETGR
MUL_0199|M.ulcerans_Agy99 TNAVTGKLAGYRVIDEQTASVTISVTRSDPSRPVDCIVRVRAKDGSETGR
MLBr_02392|M.leprae_Br4923 TSDVAGSLASYRVLDDETVSVTISVMRSDPSRPVDCIVRVRAKDGSETGR
MAV_1205|M.avium_104 TSDVKATMVGYRVLDDQTASITISVTRSDPSRPVDCIVRVRSHDGGETGR
. * . : .*.::* *..:* .* * **::*. **:*.*: :*.****
MAB_1202c|M.abscessus_ATCC_199 REVLVPPSAERSVPVTALVKTTRPPVTGDVYGCSLAIPSYLVAS--
MSMEG_5262|M.smegmatis_MC2_155 REILVPPGSDRVVQVTAEVKSTRKPVMGDIYGCGTDVPSYLVTPQT
Mflv_2049|M.gilvum_PYR-GCK RELLVPPSSETTVQVRTTVKSSRPAVIGDIYGCGIDVPDYLVTG--
TH_2168|M.thermoresistible__bu REVLVEPSVEPIVQVTTLVETSQPPLVGDIYGCGAEVPEYLIAPDR
Mb1110c|M.bovis_AF2122/97 RELLVPPSEATTVQVTTTVKSSQPPVMADVYGCGTEVPSYLRLP--
Rv1081c|M.tuberculosis_H37Rv RELLVPPSEATTVQVTTTVKSSQPPVMADVYGCGTEVPSYLRLP--
MMAR_4386|M.marinum_M REVLVPPSDQTTVQVTTTLKSSKPPLMADVYGCGTDVPAYLRSP--
MUL_0199|M.ulcerans_Agy99 REVLVPPSDQTTVQVTTTLKSSKPPLMADVYGCGTDVPAYLRSP--
MLBr_02392|M.leprae_Br4923 REVLVAPAEATTVQVVTTVKSARPPVMADIYGCGTDVPGYLRPA--
MAV_1205|M.avium_104 RELLVPPSGAATVEVTTTVKSFQPPVVADVYGCGTDVPGYLRPS--
**:** *. * * : ::: : .: .*:***. :* **