For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TEKASQSAGDPILDIDELRKEIDRLDAEILAAIKRRTEVSREIGKARMASGGPRLVHSREMKVLERYSEL GQEGHTLAMLLLRLGRGRLGH*
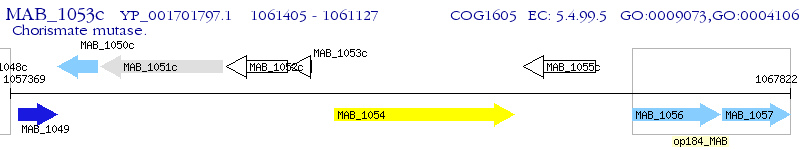
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1053c | - | - | 100% (92) | hypothetical protein MAB_1053c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0973c | - | 1e-31 | 79.01% (81) | hypothetical protein Mb0973c |
| M. gilvum PYR-GCK | Mflv_1860 | - | 2e-32 | 75.00% (92) | chorismate mutase |
| M. tuberculosis H37Rv | Rv0948c | - | 1e-31 | 79.01% (81) | hypothetical protein Rv0948c |
| M. leprae Br4923 | MLBr_00151 | - | 6e-32 | 70.97% (93) | hypothetical protein MLBr_00151 |
| M. marinum M | MMAR_4555 | - | 5e-31 | 83.12% (77) | hypothetical protein MMAR_4555 |
| M. avium 104 | MAV_1069 | - | 7e-33 | 84.62% (78) | hypothetical protein MAV_1069 |
| M. smegmatis MC2 155 | MSMEG_5536 | - | 1e-28 | 74.70% (83) | chorismate mutase |
| M. thermoresistible (build 8) | TH_1217 | - | 2e-31 | 74.71% (87) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4413 | - | 3e-31 | 83.12% (77) | hypothetical protein MUL_4413 |
| M. vanbaalenii PYR-1 | Mvan_4874 | - | 1e-30 | 74.16% (89) | chorismate mutase |
CLUSTAL 2.0.9 multiple sequence alignment
TH_1217|M.thermoresistible__bu MTT------------ETQADSDTS--VDIDALRREIDELDAQILEAVQRR
Mvan_4874|M.vanbaalenii_PYR-1 MTTM-----------ETTGSSGSSEANDIDELREEIDRLDAAILEAVQRR
Mflv_1860|M.gilvum_PYR-GCK MTT-------------THSGMETTQLPDIDDLRQEIDRLDAEILEAVKRR
MAB_1053c|M.abscessus_ATCC_199 MTE-------------KASQSAGDPILDIDELRKEIDRLDAEILAAIKRR
MSMEG_5536|M.smegmatis_MC2_155 MPE------------------TIDAVPEIDDLRREIDELDATIIAAIQRR
Mb0973c|M.bovis_AF2122/97 MRPEPPHHENAELAAMNLEMLESQPVPEIDTLREEIDRLDAEILALVKRR
Rv0948c|M.tuberculosis_H37Rv MRPEPPHHENAELAAMNLEMLESQPVPEIDTLREEIDRLDAEILALVKRR
MMAR_4555|M.marinum_M MRPEPPPHENAEL-EMNTETIDTEDLPSIDELREEIDRLDREILAAVKRR
MUL_4413|M.ulcerans_Agy99 MRPEPPPHENAEL-EMNTETIDTEDLPSIDELREEIDRLDREILAAVKRR
MLBr_00151|M.leprae_Br4923 MRPEPPHHENAELTEMNTEVVEAPLLTDIEELREEIDRLDAQILATVKRR
MAV_1069|M.avium_104 ---------------MNTETVDPREATDIDQLRREIDRLDAEILAAVKRR
.*: **.***.** *: ::**
TH_1217|M.thermoresistible__bu TEVSRLIGKTRMASGGTRLVHSREMKVIERYS-VLGPEGRELAMLLLRLG
Mvan_4874|M.vanbaalenii_PYR-1 TEVSQLIGKARMASGGTRLVHSREMKVIERYS-VLGDEGKNLAMLLLRLG
Mflv_1860|M.gilvum_PYR-GCK TEVSQMIGKARMASGGTRLVHSREMKVIERYS-VLGEEGKNLAMLLLRLG
MAB_1053c|M.abscessus_ATCC_199 TEVSREIGKARMASGGPRLVHSREMKVLERYS-ELGQEGHTLAMLLLRLG
MSMEG_5536|M.smegmatis_MC2_155 TEVSKTIGKARMASGGTRLVHSREMKVIERYIDALGPEGKDLAMLLLRLG
Mb0973c|M.bovis_AF2122/97 AEVSKAIGKARMASGGTRLVHSREMKVIERYS-ELGPDGKDLAILLLRLG
Rv0948c|M.tuberculosis_H37Rv AEVSKAIGKARMASGGTRLVHSREMKVIERYS-ELGPDGKDLAILLLRLG
MMAR_4555|M.marinum_M AEVSQAIGKARMASGGTRLVHNREMKVIERYS-ELGPEGKDLAILLLRLG
MUL_4413|M.ulcerans_Agy99 AEVSQAIGKARMASGGTRLVHNREMKVIERYS-ELGPEGKDLAILLLRLG
MLBr_00151|M.leprae_Br4923 AEVSQAIGKVRMASGGTRLVHSREMKVIERYS-ELGPDGKDLAILLLRLG
MAV_1069|M.avium_104 TEVSQAIGRARMASGGTRLVHSREMKVIERYS-ELGPDGKDLAMLLLRLG
:***: **:.******.****.*****:*** ** :*: **:******
TH_1217|M.thermoresistible__bu RGRLGH
Mvan_4874|M.vanbaalenii_PYR-1 RGRLGK
Mflv_1860|M.gilvum_PYR-GCK RGRLGH
MAB_1053c|M.abscessus_ATCC_199 RGRLGH
MSMEG_5536|M.smegmatis_MC2_155 RGRLGY
Mb0973c|M.bovis_AF2122/97 RGRLGH
Rv0948c|M.tuberculosis_H37Rv RGRLGH
MMAR_4555|M.marinum_M RGRLGH
MUL_4413|M.ulcerans_Agy99 RGRLGH
MLBr_00151|M.leprae_Br4923 RGRLGH
MAV_1069|M.avium_104 RGRLGH
*****