For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLAAAWQALETHHKQIASTHLREFFAEDPARGSELTVTVGDLYIDYSKHRLNRETLALLVDLAKAADLQG RRDAMFAGAHINTSEDRAVLHTALRLPRDAKLVVDGQDVVADVHQVLDAMGDFTDRLRSGEWTGATGKRI TTVVNIGIGGSDLGPVMVYQALRHYADAGISARFVSNVDPADLVATLADLDPATTLFIVASKTFSTLETL TNATAARRWLVAALGDEAVSKHFVAVSTNAKLVSEFGIDTANMFGFWDWVGGRYSVDSAIGLSVMAVIGR EAFGQFLAGFHIVDEHFRTAPLEQNAPALLGLIGLWYSNFFGAQSRGVLPYSNDLSRFAAYLQQLTMESN GKSVKADGAPVSVDTGDIYWGEPGTNGQHAFYQLLHQGTRLIPADFIGFSEPTDDLPTADGTGSMHDLLM SNFFAQTQVLAFGKTAEEITAEGTAPNVVPHKVMPGNRPSTTILASKLTPSTVGQLIALYEHQVFTAGTV WGIDSFDQWGVELGKKQAIALLPVITDDTSPAQQTDSSTDTLVRHYRNARGRTT*
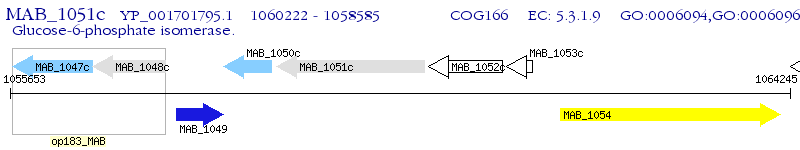
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1051c | pgi | - | 100% (545) | glucose-6-phosphate isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0971c | pgi | 0.0 | 83.79% (543) | glucose-6-phosphate isomerase |
| M. gilvum PYR-GCK | Mflv_1857 | pgi | 0.0 | 82.35% (544) | glucose-6-phosphate isomerase |
| M. tuberculosis H37Rv | Rv0946c | pgi | 0.0 | 83.79% (543) | glucose-6-phosphate isomerase |
| M. leprae Br4923 | MLBr_00150 | pgi | 0.0 | 80.18% (545) | glucose-6-phosphate isomerase |
| M. marinum M | MMAR_4557 | pgi | 0.0 | 83.82% (544) | glucose-6-phosphate isomerase Pgi |
| M. avium 104 | MAV_1068 | pgi | 0.0 | 83.30% (545) | glucose-6-phosphate isomerase |
| M. smegmatis MC2 155 | MSMEG_5541 | pgi | 0.0 | 84.90% (543) | glucose-6-phosphate isomerase |
| M. thermoresistible (build 8) | TH_1684 | pgi | 1e-170 | 57.57% (535) | PROBABLE GLUCOSE-6-PHOSPHATE ISOMERASE PGI (GPI) |
| M. ulcerans Agy99 | MUL_4416 | pgi | 0.0 | 83.09% (544) | glucose-6-phosphate isomerase |
| M. vanbaalenii PYR-1 | Mvan_4877 | pgi | 0.0 | 83.21% (548) | glucose-6-phosphate isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4557|M.marinum_M MTSEQSIPDITATPAWEALRKHHDQIGETHLRQIFADDPNRGHDLTVTVG
MUL_4416|M.ulcerans_Agy99 MTSEQPIPDITATPAWEALRKHHDQIGETHLRQIFADDPNRGHDLTVTVG
MLBr_00150|M.leprae_Br4923 MTSMQAIPDITATPAWDALRRHHDEIGATHLRQFFADNPNRGRELVITVG
MAV_1068|M.avium_104 MTSVHTLPDITATPAWDALRKHHDRIGDTHLRQFFEEDPDRGRELTVTVG
Mb0971c|M.bovis_AF2122/97 MTS-APIPDITATPAWDALRRHHDQIGNTHLRQFFADDPGRGRELTVSVG
Rv0946c|M.tuberculosis_H37Rv MTS-APIPDITATPAWDALRRHHDQIGNTHLRQFFADDPGRGRELTVSVG
Mflv_1857|M.gilvum_PYR-GCK MGT-----DITATSEWQALARHRDEIGETNLRQLFADDAARGLRFAVTVG
Mvan_4877|M.vanbaalenii_PYR-1 MGS-----DITATPAWQALSRHHDEIRAKDLRGLFADDPTRGTGFALTVG
MSMEG_5541|M.smegmatis_MC2_155 MSA-----DITETPAWQALSDHHAEIGDRHLTELFADDPARGTELALTVG
MAB_1051c|M.abscessus_ATCC_199 ---------MTLAAAWQALETHHKQIASTHLREFFAEDPARGSELTVTVG
TH_1684|M.thermoresistible__bu ------------------LRTHREHLSDVSIAELFAADPDRGPAMTVEHD
* *: .: : :* :. ** :.: .
MMAR_4557|M.marinum_M DLYIDYSKHRITRDTISLLVDLARTANLEAHRDQMFAGAHINTSEDRAVL
MUL_4416|M.ulcerans_Agy99 DLYIDYSKHRITRDTISLLVDLARTANLEAHRDQMFAGAHINTSEDRAVL
MLBr_00150|M.leprae_Br4923 DLYIDYSKHRITHDTVQLLVDLARAANLEQRRDQMLAGVHVNTSENRSVL
MAV_1068|M.avium_104 DLYIDYSKHRVTRETLRLLVDLARTAKLEERRDQMFAGVHINTSEDRAVL
Mb0971c|M.bovis_AF2122/97 DLYIDYSKHRVTRETLALLIDLARTAHLEERRDQMFAGVHINTSEDRAVL
Rv0946c|M.tuberculosis_H37Rv DLYIDYSKHRVTRETLALLIDLARTAHLEERRDQMFAGVHINTSEDRAVL
Mflv_1857|M.gilvum_PYR-GCK GLYIDYSKHRVTAETLNLLVDVARAAGLETKRDAMFSGEHINTSEDRAVL
Mvan_4877|M.vanbaalenii_PYR-1 DLYIDYSKHRVTRETLDLLLDLARTAGLPAKRDAMFSGAHINTSEDRAVL
MSMEG_5541|M.smegmatis_MC2_155 DLYIDYSKHRVTRRTLDLLVDLARAAGLEERRDAMFAGEHINTSEDRAVL
MAB_1051c|M.abscessus_ATCC_199 DLYIDYSKHRLNRETLALLVDLAKAADLQGRRDAMFAGAHINTSEDRAVL
TH_1684|M.thermoresistible__bu GIVLDYSKNLLTARTVELLAEFAAERDLDSGLTALFAGDRINTSENRPAL
.: :****: :. *: ** :.* * :::* ::****:*..*
MMAR_4557|M.marinum_M HTALRLPRDAELIVDGRNVVEDVHAVLDAMGDFTDRLRSGEWTGATGKRI
MUL_4416|M.ulcerans_Agy99 HTALRLPRDAELIVDGRNVVEDVHAVLDAMGDFTDRLRSGEWTGATGKRI
MLBr_00150|M.leprae_Br4923 HTALRLPRDTELIVDGQNVVQDVHAVLDVMGDFTDRLRSGEWTGATGKRI
MAV_1068|M.avium_104 HTALRLPRDAELIVDGRNVVADVHEVLDAMGEFTDRLRSGEWTGATGKRI
Mb0971c|M.bovis_AF2122/97 HTALRLPRDAELVVDGQDVVTDVHAVLDAMGAFTDRLRSGEWTGATGKRI
Rv0946c|M.tuberculosis_H37Rv HTALRLPRDAELVVDGQDVVTDVHAVLDAMGAFTDRLRSGEWTGATGKRI
Mflv_1857|M.gilvum_PYR-GCK HTALRLPRDAALTVDGQDVVADVHEVLDRMGDFTDRLRSGDWLGATGERI
Mvan_4877|M.vanbaalenii_PYR-1 HTALRLPRDASLTVDGQDVVADVHEVLDRMGDFTDRLRSGEWRGATGERI
MSMEG_5541|M.smegmatis_MC2_155 HTALRLPRDAKLVVDGQDVVADVHDVLDRMGDFTDRLRSGEWTGATGERI
MAB_1051c|M.abscessus_ATCC_199 HTALRLPRDAKLVVDGQDVVADVHQVLDAMGDFTDRLRSGEWTGATGKRI
TH_1684|M.thermoresistible__bu HTALRAPADDEIIVDGSDVVPAVHDVLARMGEFATRVRDGAWRGATGDAI
***** * * : *** :** ** ** ** *: *:*.* * ****. *
MMAR_4557|M.marinum_M STVVNIGIGGSDLGPVMVYQALRHYADAGISARFVSNVDPADLIATLADL
MUL_4416|M.ulcerans_Agy99 STVVNIGIGGSDLGPVMVYQALRHYADAGISARFVSNVDPADLIATLADL
MLBr_00150|M.leprae_Br4923 NTVVNIGIGGSDLGPVMVYQALRHYADAGISARFVSNIDPADLTAKLSDL
MAV_1068|M.avium_104 STVVNIGIGGSDLGPVMVYQALRHYADAGISARFVSNVDPADLIATLADL
Mb0971c|M.bovis_AF2122/97 STVVNIGIGGSDLGPVMVYQALRHYADAGISARFVSNVDPADLIATLADL
Rv0946c|M.tuberculosis_H37Rv STVVNIGIGGSDLGPVMVYQALRHYADAGISARFVSNVDPADLIATLADL
Mflv_1857|M.gilvum_PYR-GCK TTVVNIGIGGSDLGPVMVDQALRHYADAGISARFVSNVDPADLVAKLAGL
Mvan_4877|M.vanbaalenii_PYR-1 TTVVNIGIGGSDLGPVMVDQALRHYADAGISARFVSNVDPADLVAKLNGL
MSMEG_5541|M.smegmatis_MC2_155 TTVVNIGIGGSDLGPVMVYDALRHYADAGISARFVSNVDPADLVAKLDGL
MAB_1051c|M.abscessus_ATCC_199 TTVVNIGIGGSDLGPVMVYQALRHYADAGISARFVSNVDPADLVATLADL
TH_1684|M.thermoresistible__bu RTVVNIGIGGSDLGPAMAYQALRHAAELDV--RFVANVDGHDLAGVLRSL
**************.*. :**** *: .: ***:*:* ** . * .*
MMAR_4557|M.marinum_M DPATTLFIVASKTFSTLETLTNATAARRWLTDALG----DAAVSQHFVAV
MUL_4416|M.ulcerans_Agy99 DPATTLFIVASKTFSTLETLTNATAARRWLTDALG----DAAVSQHFVAV
MLBr_00150|M.leprae_Br4923 EPGTTLFIVASKTFSTLETLTNATAARRWLTDALG----EAAVSKHFVAV
MAV_1068|M.avium_104 DPATTLFIVASKTFSTLETLTNATAARRWITDALG----DAAVAHHFVAV
Mb0971c|M.bovis_AF2122/97 DPATTLFIVASKTFSTLETLTNATAARRWLTDALG----DAAVSRHFVAV
Rv0946c|M.tuberculosis_H37Rv DPATTLFIVASKTFSTLETLTNATAARRWLTDALG----DAAVSRHFVAV
Mflv_1857|M.gilvum_PYR-GCK NPATTLFVIASKTFSTLETLTNATAARRWLIEALG----EDAVSKHFVAV
Mvan_4877|M.vanbaalenii_PYR-1 DPATTLFIIASKTFSTLETLTNATAARRWLVDGLGGSAGQDAVSKHFVAV
MSMEG_5541|M.smegmatis_MC2_155 EPAKTLFIVASKTFSTLETLTNATAARRWLTDALG----DAAVAKHFVAV
MAB_1051c|M.abscessus_ATCC_199 DPATTLFIVASKTFSTLETLTNATAARRWLVAALG----DEAVSKHFVAV
TH_1684|M.thermoresistible__bu DPATTLFIVSSKTFTTTETLLNAHSARTWLQSALG----DDAVSRHFVAV
:*..***:::****:* *** ** :** *: .** : **::*****
MMAR_4557|M.marinum_M STNRRLVDDFGINTDNMFGFWDWVGGRYSVDSAIGLSVMAAIGREAFADF
MUL_4416|M.ulcerans_Agy99 STNRRLVDDFGINTDNIFGFWDWVGGRYSVDSAIGLSVMAAIGREAFADF
MLBr_00150|M.leprae_Br4923 STNKRLVKDFGINTANMFGFWEWVGGRYSVDSAIGLSLMAVVGRESFADF
MAV_1068|M.avium_104 STNKRLVDDFGINTDNMFGFWDWVGGRYSVDSAIGLSVMAVIGREAFADF
Mb0971c|M.bovis_AF2122/97 STNKRLVDDFGINTDNMFGFWDWVGGRYSVDSAIGLSLMTVIGRDAFADF
Rv0946c|M.tuberculosis_H37Rv STNKRLVDDFGINTDNMFGFWDWVGGRYSVDSAIGLSLMTVIGRDAFADF
Mflv_1857|M.gilvum_PYR-GCK STNAKLVAEFGIDTDNMFGFWDWVGGRYSVDSAIGLSVMAVIGRERFAEF
Mvan_4877|M.vanbaalenii_PYR-1 STNAKLVDDFGIDTANMFGFWDWVGGRYSVDSAIGLSVMAVIGRERFAEF
MSMEG_5541|M.smegmatis_MC2_155 STNKKLVDEFGINTDNMFGFWDWVGGRYSVDSAIGLSVMAVIGKERFAEF
MAB_1051c|M.abscessus_ATCC_199 STNAKLVSEFGIDTANMFGFWDWVGGRYSVDSAIGLSVMAVIGREAFGQF
TH_1684|M.thermoresistible__bu STNTRAVTEFGITADNMFEFWDWVGGRYSIHSAIGLSLMLAIGPGRFREF
*** : * :*** : *:* **:*******:.******:* .:* * :*
MMAR_4557|M.marinum_M LSGFHIVDEHFRTAPLESNAPALLGLIGLWYSNFLGAQSRAVLPYSNDLA
MUL_4416|M.ulcerans_Agy99 LSGFHIVDEHFRTAPPESNAPALLGLIGLWYSNFLGAQSRAVLPYSNDLA
MLBr_00150|M.leprae_Br4923 LSGFHIVDQHFQNAPLESNAPVLLGLIGLWYSDFLGAQSRAVLPYSNDLA
MAV_1068|M.avium_104 LSGFHIVDRHFQTAPLESNAPVLLGLIGLWYSNFMGAQSRAVLPYSNDLA
Mb0971c|M.bovis_AF2122/97 LAGFHIIDRHFATAPLESNAPVLLGLIGLWYSNFFGAQSRTVLPYSNDLS
Rv0946c|M.tuberculosis_H37Rv LAGFHIIDRHFATAPLESNAPVLLGLIGLWYSNFFGAQSRTVLPYSNDLS
Mflv_1857|M.gilvum_PYR-GCK LAGFHLIDEHFRTAPLEANAPVLLGLIGLWYNNFFGAETRAVLPYSNDLA
Mvan_4877|M.vanbaalenii_PYR-1 LAGFHLVDEHFRSAPLEANAPVLLGLIGLWYSNFFGAETRAVLPYSNDLA
MSMEG_5541|M.smegmatis_MC2_155 LAGFHIVDEHFRTAPLHQNAPALLGLIGLWYSNFFGAQSRAVLPYSNDLS
MAB_1051c|M.abscessus_ATCC_199 LAGFHIVDEHFRTAPLEQNAPALLGLIGLWYSNFFGAQSRGVLPYSNDLS
TH_1684|M.thermoresistible__bu LAGAHSMDRHFRTAPVLQNIPILLALLGIWYRNFWGAQTHAVLPYDQRLS
*:* * :*.** .** * * **.*:*:** :* **::: ****.: *:
MMAR_4557|M.marinum_M RFAAYLQQLTMESNGKSTRADGTAVTTDTGEIYWGEPGTNGQHAFYQLLH
MUL_4416|M.ulcerans_Agy99 RFAAYLQQLTMESNGKSTRADGTAVTTDTGEIYWGERGTNGQHAFYQLLH
MLBr_00150|M.leprae_Br4923 RFAAYLQQLTMESNGKSTRADGTPVTTNTGEIYWGETGTNGQHAFYQLLH
MAV_1068|M.avium_104 RFAAYLQQLTMESNGKSTRADGSPVTTDTGEIFWGEPGTNGQHAFYQLLH
Mb0971c|M.bovis_AF2122/97 RFPAYLQQLTMESNGKSTRADGSPVSADTGEIFWGEPGTNGQHAFYQLLH
Rv0946c|M.tuberculosis_H37Rv RFPAYLQQLTMESNGKSTRADGSPVSADTGEIFWGEPGTNGQHAFYQLLH
Mflv_1857|M.gilvum_PYR-GCK RFAAYLQQLTMESNGKSVQADGTPVSTATGEIFWGEPGTNGQHAFYQLLH
Mvan_4877|M.vanbaalenii_PYR-1 RFAAYLQQLTMESNGKSVRADGTPVSTGTGEIFWGEPGTNGQHAFYQLLH
MSMEG_5541|M.smegmatis_MC2_155 RFAAYLQQLTMESNGKSVRADGTPVSTDTGEIFWGEPGTNGQHAFYQLLH
MAB_1051c|M.abscessus_ATCC_199 RFAAYLQQLTMESNGKSVKADGAPVSVDTGDIYWGEPGTNGQHAFYQLLH
TH_1684|M.thermoresistible__bu RLPAYLQQLEMESNGKSVTRDGHGCAIPTAPVIWGEPGTNGQHAFFQLLH
*:.****** *******. ** : *. : *** ********:****
MMAR_4557|M.marinum_M QGTRLVPADFIGFSQPIDDLPTVEGTGSMHDLLMSNFFAQTQVLAFGKTA
MUL_4416|M.ulcerans_Agy99 QGTRLVPADFIGFSQPIDDLPTVEGTGSMHDLLMSNFFAQTQVLAFGKTA
MLBr_00150|M.leprae_Br4923 QGTRLVPADFIGFSQPIDDLPTVDGIGSMHDLLMSNFFAQTQVLAFGKTA
MAV_1068|M.avium_104 QGTRLVPADFIGFSQPIDDLPTAEGSGSMHDLLMSNFFAQTQVLAFGKTA
Mb0971c|M.bovis_AF2122/97 QGTRLVPADFIGFAQPLDDLPTAEGTGSMHDLLMSNFFAQTQVLAFGKTA
Rv0946c|M.tuberculosis_H37Rv QGTRLVPADFIGFAQPLDDLPTAEGTGSMHDLLMSNFFAQTQVLAFGKTA
Mflv_1857|M.gilvum_PYR-GCK QGTRLVPADFIGFSEPTDDLPTADGTGSMHDLLMSNFFAQTQVLAFGKTA
Mvan_4877|M.vanbaalenii_PYR-1 QGTRLVPADFIGFSEPTDDLPTADGTGSMHDLLMSNFFAQTQVLAFGKTA
MSMEG_5541|M.smegmatis_MC2_155 QGTRLVPADFIGFSQPTDDLPTADGTGSMHDLLMSNFFAQTQVLAFGKTA
MAB_1051c|M.abscessus_ATCC_199 QGTRLIPADFIGFSEPTDDLPTADGTGSMHDLLMSNFFAQTQVLAFGKTA
TH_1684|M.thermoresistible__bu QGSVLVPCDFIGVLRPGHDL------AGHHDVLMANLFAQTEALAFGRPQ
**: *:*.****. .* .** .. **:**:*:****:.****:.
MMAR_4557|M.marinum_M EEIAAEGTPAAVVPHKVMPGNRPSTSILANRLTPSVLGQLIALYEHQVFT
MUL_4416|M.ulcerans_Agy99 EEIAAEGTPAAVVPHKVMPGNRPSTSILAKRLTPSVLGQLIAVYEHQVFT
MLBr_00150|M.leprae_Br4923 EEIAAEGTPAEVVPHKVMPGNRPTTSILANRLTPSVLGQLIALYEHQVFT
MAV_1068|M.avium_104 EEIAAEGTPADIVPHKVMPGNRPSTSILANRLTPSVLGQLIALYEHQVFT
Mb0971c|M.bovis_AF2122/97 EEIAADGTPAHVVAHKVMPGNRPSTSILASRLTPSVLGQLIALYEHQVFT
Rv0946c|M.tuberculosis_H37Rv EEIAADGTPAHVVAHKVMPGNRPSTSILASRLTPSVLGQLIALYEHQVFT
Mflv_1857|M.gilvum_PYR-GCK DEISAEGTSPDVVPHKVMPGNRPTTSILATKLTPSVVGQLIALYEHQVFV
Mvan_4877|M.vanbaalenii_PYR-1 EEITGEGTPPNVVPHKVMPGNRPSTSILATKLTPSVVGQLIALYEHQVFV
MSMEG_5541|M.smegmatis_MC2_155 DAIASEGTPADVVPHKVMPGNRPTTSILATKLTPSVVGQLIALYEHQVFT
MAB_1051c|M.abscessus_ATCC_199 EEITAEGTAPNVVPHKVMPGNRPSTTILASKLTPSTVGQLIALYEHQVFT
TH_1684|M.thermoresistible__bu PDNGPAP------AHRRFPGNRPSSTILLPSLTPYTLGQLIALYEHKVFT
.*: :*****:::** *** .:*****:***:**.
MMAR_4557|M.marinum_M EGVVWGIDSFDQWGVELGKTQAKALLPVITSDGSPQRQSDSSTDALVRRY
MUL_4416|M.ulcerans_Agy99 EGVVWGIDSFDQWGVELGKTQAKALLPVITSDGSPQRQSDSSTDALVRRY
MLBr_00150|M.leprae_Br4923 EGVIWGIDSFDQWGVELGKKQAEALLPVITGNASPAQQLDSSTDTLVRRY
MAV_1068|M.avium_104 EGVIWGIDSFDQWGVELGKTQAKALLPVITADNSPAPQSDSSTDALVRRY
Mb0971c|M.bovis_AF2122/97 EGVVWGIDSFDQWGVELGKTQAKALLPVITGAGSPPPQSDSSTDGLVRRY
Rv0946c|M.tuberculosis_H37Rv EGVVWGIDSFDQWGVELGKTQAKALLPVITGAGSPPPQSDSSTDGLVRRY
Mflv_1857|M.gilvum_PYR-GCK EGVIWGIDSFDQWGVELGKTQAKALLPVLTDAEPPAAQSDSSTDALVRRY
Mvan_4877|M.vanbaalenii_PYR-1 EGVIWGIDSFDQWGVELGKTQAKALLPVLTGADSPAAQSDSSTDALVRRY
MSMEG_5541|M.smegmatis_MC2_155 EGVIWGIDSFDQWGVELGKTQAKALLPVLTGDKSPAAQSDTSTDALVRRY
MAB_1051c|M.abscessus_ATCC_199 AGTVWGIDSFDQWGVELGKKQAIALLPVITDDTSPAQQTDSSTDTLVRHY
TH_1684|M.thermoresistible__bu QGWLWGINSFDQWGVELGKVLAARITTEFGPDAG--HAHDSSTNALIARY
* :***:*********** * : . : *:**: *: :*
MMAR_4557|M.marinum_M RTQRGRTG---
MUL_4416|M.ulcerans_Agy99 RTQRGRTG---
MLBr_00150|M.leprae_Br4923 RTERGRTS---
MAV_1068|M.avium_104 RSERGRTS---
Mb0971c|M.bovis_AF2122/97 RTERGRAG---
Rv0946c|M.tuberculosis_H37Rv RTERGRAG---
Mflv_1857|M.gilvum_PYR-GCK RSERGRTA---
Mvan_4877|M.vanbaalenii_PYR-1 RTERGRTA---
MSMEG_5541|M.smegmatis_MC2_155 RTERGRPA---
MAB_1051c|M.abscessus_ATCC_199 RNARGRTT---
TH_1684|M.thermoresistible__bu RKARSETAGQA
*. *...