For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLIDLTPEQAKLQNDLRHYFSNLVTAEETREMMIDRHGSSYEAVVRRMGQDGWLGVGWPKEYGGHGFGPL EQQIFMNEVMRADVPMPLVTLQTVGPTLQKYGTEEQKKKFLPGILAGEIHFAIGYTEPEAGTDLASLRTT AVRDGDHYVVNGQKIFTTGAHQAQYIWLACRTDPSAPKHKGISILIVDTKDPGYSWTPIITNDGAHHTNA TYYTDVRVPVEMLVGEENAGWKLITTQLNHERVSLGPSGRIAGMYDRVYEWASKPGPDGVAPLSHEDVRR VLGEMKAVWRLNELLNWQVASSGDDISMADAAATKILATEWIQKLGRLSEGVVGRYGDPADANTAETLEW LDAQTKRHLVITFGGGVNEVMREMVSTAGLGTPRVPR
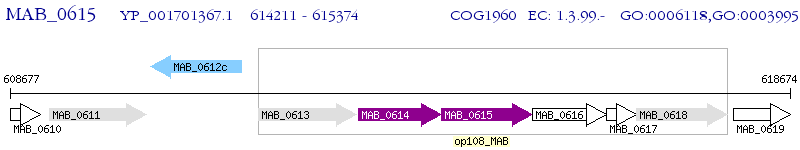
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0615 | - | - | 100% (387) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4158 | - | 2e-90 | 46.06% (393) | acyl-CoA dehydrogenase FadE |
| M. abscessus ATCC 19977 | MAB_0597 | - | 2e-44 | 31.12% (392) | acyl-CoA dehydrogenase FadE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3573c | fadE29 | 1e-180 | 75.19% (387) | acyl-CoA dehydrogenase FADE29 |
| M. gilvum PYR-GCK | Mflv_1510 | - | 1e-177 | 74.16% (387) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3543c | fadE29 | 1e-179 | 75.19% (387) | acyl-CoA dehydrogenase FADE29 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 6e-24 | 25.77% (357) | putative acyl-CoA dehydrogenase |
| M. marinum M | MMAR_5030 | fadE29 | 1e-178 | 74.68% (387) | acyl-CoA dehydrogenase FadE29 |
| M. avium 104 | MAV_0618 | - | 1e-179 | 75.45% (387) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5993 | - | 1e-177 | 72.61% (387) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4261 | - | 1e-177 | 73.66% (391) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_4104 | fadE29 | 1e-177 | 74.16% (387) | acyl-CoA dehydrogenase FadE29 |
| M. vanbaalenii PYR-1 | Mvan_5256 | - | 1e-179 | 74.68% (387) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5993|M.smegmatis_MC2_155 ---------------MFIDLTPEQRKLQSELREYFSTLLTPEEAAAMETD
Mvan_5256|M.vanbaalenii_PYR-1 ---------------MYIDLTPEQRQLQAELRQYFSTLITPDEAAAMETD
Mflv_1510|M.gilvum_PYR-GCK ---------------MYIELTPEQRSLQAELREYFTTLITPEEAEAMKSD
Mb3573c|M.bovis_AF2122/97 ---------------MFIDLTPEQRQLQAEIRQYFSNLISPDERTEMEKD
Rv3543c|M.tuberculosis_H37Rv ---------------MFIDLTPEQRQLQAEIRQYFSNLISPDERTEMEKD
MMAR_5030|M.marinum_M ---------------MFIDLTPEQRQLQAELRQYFSNLISSDEMKAMEKD
MUL_4104|M.ulcerans_Agy99 ---------------MFIDLTPEQRQLQAELRQYFSNLISSDEMKAMEKD
MAV_0618|M.avium_104 ---------------MFIDLTPEQRQLQAELREYFSNLISSDEMKAMEQD
TH_4261|M.thermoresistible__bu MPPWAAGGQGTTGTEMYIELTPEQKKLQAELREYFANLISPEERRAMEVD
MAB_0615|M.abscessus_ATCC_1997 ---------------MLIDLTPEQAKLQNDLRHYFSNLVTAEETREMMID
MLBr_00737|M.leprae_Br4923 -----MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAP---HAADVD
:.*. *: .*: :* . ::. *
MSMEG_5993|M.smegmatis_MC2_155 RHNEAYRAVIKRMGADGKLGVGWPKEYGGLGYGPVEQQIFINEANRADIP
Mvan_5256|M.vanbaalenii_PYR-1 RHNEAYRAVIKRMGSDGKLGVGWPKEYGGLGFGPVEQQIFVNEANRADVP
Mflv_1510|M.gilvum_PYR-GCK RHNEAYRAVIKRMGSDGKLGVGWPKEYGGLGFGPIEQQIFVNEANRADVP
Mb3573c|M.bovis_AF2122/97 RHGPAYRAVIRRMGRDGRLGVGWPKEFGGLGFGPIEQQIFVNEAHRADVP
Rv3543c|M.tuberculosis_H37Rv RHGPAYRAVIRRMGRDGRLGVGWPKEFGGLGFGPIEQQIFVNEAHRADVP
MMAR_5030|M.marinum_M RHNAAYRAVIRRMGKDGKLGVGWPKEFGGLGFGPIEQQIFVNEAHRADVP
MUL_4104|M.ulcerans_Agy99 RHNAAYRAVIRRMGKDGKLGVGWPKEFGGLGFGPIEQQIFVNEAHRADVP
MAV_0618|M.avium_104 RHNEAYRAVIRRMGRDGKLGVGWPKEFGGLGFGPIEQSIFVNEAHRADVP
TH_4261|M.thermoresistible__bu RHNEAYRAVIRRMGADGKLGVGWPKEFGGLGYGPVEQSIFVNEAARADVP
MAB_0615|M.abscessus_ATCC_1997 RHGSSYEAVVRRMGQDGWLGVGWPKEYGGHGFGPLEQQIFMNEVMRADVP
MLBr_00737|M.leprae_Br4923 QRARFPEEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDAS
:: . .: :. .* .: *:*:** * ..: *.::*. *.* .
MSMEG_5993|M.smegmatis_MC2_155 LPMV-TLQTVGPT-LQVYGTEEQKKKFLPGILSGDVHFAIGYSEPEAGTD
Mvan_5256|M.vanbaalenii_PYR-1 LPLV-TLQTVGPT-LQVYGTEEQKKKFLPGILSGDIHFAIGYSEPDAGTD
Mflv_1510|M.gilvum_PYR-GCK LPMV-TLQTVGPT-LQVHGTEEQKKKFLPGILSGEVHFAIGYSEPEAGTD
Mb3573c|M.bovis_AF2122/97 LPAV-TLQTVGPT-LQAHGSELQKKKFLPAILAGEAHFAIGYTEPEAGTD
Rv3543c|M.tuberculosis_H37Rv LPAV-TLQTVGPT-LQAHGSELQKKKFLPAILAGEAHFAIGYTEPEAGTD
MMAR_5030|M.marinum_M LPAV-TLQTVGPT-LQAYGSDAQKKKFLPAILAGEVHFAIGYTEPEAGTD
MUL_4104|M.ulcerans_Agy99 LPAV-TLQTVGPT-LQAYGSDAQKKKFLPAILAGEVHFAIGYTEPEAGTD
MAV_0618|M.avium_104 LPAV-TLQTVGPT-LQQFGSEMQKKKFLPAILAGEVHFAIGYTEPEAGTD
TH_4261|M.thermoresistible__bu LPAV-TLQTVGPT-LQVYGTEEQKKKFLPAILAGEVHFAIGYTEPEAGTD
MAB_0615|M.abscessus_ATCC_1997 MPLV-TLQTVGPT-LQKYGTEEQKKKFLPGILAGEIHFAIGYTEPEAGTD
MLBr_00737|M.leprae_Br4923 ASLIPAVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSD
. : :::.:*. * *:: **:.**.: : : . :* :**:*
MSMEG_5993|M.smegmatis_MC2_155 LASLRTTAIRH----GDEYIVNGQKMWTTGAHDADYIWLACRTDPEAPKH
Mvan_5256|M.vanbaalenii_PYR-1 LASLRTTAVKH----GDEYIVNGQKMWTTGAHDADYIWLACRTDPEAAKH
Mflv_1510|M.gilvum_PYR-GCK LASLRTTAVRH----GDEYVVNGQKMWTTGAHDADYIWLACRTDPEAAKH
Mb3573c|M.bovis_AF2122/97 LASLRTTAVRD----GDHYIVNGQKVFTTGAHDADYIWLACRTDPNAAKH
Rv3543c|M.tuberculosis_H37Rv LASLRTTAVRD----GDHYIVNGQKVFTTGAHDADYIWLACRTDPNAAKH
MMAR_5030|M.marinum_M LASLRTTAVRQ----GDEYIVNGQKIFTTGAHDADYIWLACRTDPEAVKH
MUL_4104|M.ulcerans_Agy99 LASLRTTAVRQ----GDEYIVNGQKIFTTGAHDADYIWLACRTDPEAVKH
MAV_0618|M.avium_104 LASLRTTAVRQ----GDEYIVNGQKIFTTGAHDADYIWLACRTDPEAVKH
TH_4261|M.thermoresistible__bu LASLRTTAVRRKEGSAEYYLVNGQKVFTTGAHDADYIWLACRTDPNAAKH
MAB_0615|M.abscessus_ATCC_1997 LASLRTTAVRD----GDHYVVNGQKIFTTGAHQAQYIWLACRTDPSAPKH
MLBr_00737|M.leprae_Br4923 AASMRTRAKAD----GDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGAN
**:** * .: :::** * : *.. .: : : . ***. :
MSMEG_5993|M.smegmatis_MC2_155 KGISILIVDTKDPGYSWTP--IILSDGAHHTNASYYNDVHVPADMLVGEE
Mvan_5256|M.vanbaalenii_PYR-1 KGISILIVDTKDPGYSWTP--IILSDGAHHTNASYYNDVRVPADMLVGEE
Mflv_1510|M.gilvum_PYR-GCK KGISILIVDTKDPGYSWTP--IILSDGAHHTNASYYNDVRVPADMLVGEE
Mb3573c|M.bovis_AF2122/97 KGISILIVDTKDPGYSWTP--IILADGAHHTNATYYNDVRVPVDMLVGKE
Rv3543c|M.tuberculosis_H37Rv KGISILIVDTKDPGYSWTP--IILADGAHHTNATYYNDVRVPVDMLVGKE
MMAR_5030|M.marinum_M KGISILIVDTKDPGYSWTP--IILSDGAHHTNATYYNDVRVPADMLVGEP
MUL_4104|M.ulcerans_Agy99 KGISILIVDTKDLGYSWAP--IILSDGAHHTNATYYNDVRVPADMLVGEP
MAV_0618|M.avium_104 KGISILIVDTKDPGYSWTP--IILSDGAHHTNATYYNDVRVPADMLVGEE
TH_4261|M.thermoresistible__bu KGISILIVDTKDPGYSWTP--MILSDGAHHTNATYYNDVRVPADMLVGEE
MAB_0615|M.abscessus_ATCC_1997 KGISILIVDTKDPGYSWTP--IITNDGAHHTNATYYTDVRVPVEMLVGEE
MLBr_00737|M.leprae_Br4923 -GISAFIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEP
*** :**...* *:* * . *. *: . ::* : ::*:
MSMEG_5993|M.smegmatis_MC2_155 NGGWKLITTQLNHERVGLGP--AGRVAGIYDQVHKWASTPGSDGVTPIEH
Mvan_5256|M.vanbaalenii_PYR-1 NGGWKLITTQLNHERVGLGP--AGRVAGIYDQVHAWASKPGSDGVTPVEH
Mflv_1510|M.gilvum_PYR-GCK NGGWKLITTQLNHERVGLGP--AGRVAGIYDEVREWATTPGSHGVVPIEQ
Mb3573c|M.bovis_AF2122/97 NDGWRLITTQLNNERVMLGP--AGRFASIYDRVHAWASVPGGNGVTPIDH
Rv3543c|M.tuberculosis_H37Rv NDGWRLITTQLNNERVMLGP--AGRFASIYDRVHAWASVPGGNGVTPIDH
MMAR_5030|M.marinum_M NGGWRLITTQLNNERVMLGP--AGRFASIYDRVRAWAAKPGGDGVTPIDH
MUL_4104|M.ulcerans_Agy99 NGGWRLITTQLNNERVMLGP--AGRFASIYDRVRAWAAKPGGDGVTPIDH
MAV_0618|M.avium_104 NGGWKLITTQLNNERVMLGP--AGRTAGIYDRVREWASKPGSDGVTPIDH
TH_4261|M.thermoresistible__bu NGGWKLITTQLNHERVMLGP--AGKVAGIYDKVHAWATKPGPSGGAPIDD
MAB_0615|M.abscessus_ATCC_1997 NAGWKLITTQLNHERVSLGP--SGRIAGMYDRVYEWASKPGPDGVAPLSH
MLBr_00737|M.leprae_Br4923 GTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTF
. *:: : *:: * :*. * . * . :: *
MSMEG_5993|M.smegmatis_MC2_155 PEAQRLLGQIKAIWRINELLNWQVAA---SGETIAVADAAATKVFSTERI
Mvan_5256|M.vanbaalenii_PYR-1 DDVKRLLGQIKAIWRVNELLNWQVAA---SGETIAVADAAATKVFSTERI
Mflv_1510|M.gilvum_PYR-GCK EGARRLLGQIKAIWRINELLNWQVAA---SGETIAVADAAATKVFSTERI
Mb3573c|M.bovis_AF2122/97 DDVKRALGEIRAIWRINELLNWQVAS---AGEDINMADAAATKVFGTERV
Rv3543c|M.tuberculosis_H37Rv DDVKRALGEIRAIWRINELLNWQVAS---AGEDINMADAAATKVFGTERV
MMAR_5030|M.marinum_M DDVRRALGEIHATWRINELLNWQVAA---AGENINVADAAATKVFGTERV
MUL_4104|M.ulcerans_Agy99 DDVRRALGEIHATWRINELLNWQVAA---AGENINVADAAATKVFGTERV
MAV_0618|M.avium_104 ADVQRALGEIYAMWRINELLNWQVAA---AGEDINVADAASTKVFGTERI
TH_4261|M.thermoresistible__bu ANVRRILGEIRAIWRINELLNWQVAA---AGETIAVADAAATKVFSTERI
MAB_0615|M.abscessus_ATCC_1997 EDVRRVLGEMKAVWRLNELLNWQVAS---SGDDISMADAAATKILATEWI
MLBr_00737|M.leprae_Br4923 QSIQFMLADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIA
: *.:: . .*: : .*: *: :*::* :.::
MSMEG_5993|M.smegmatis_MC2_155 QEVGRLAEEVIARFGNPADRHTADLMVWIDKMTKRNLVITFGGGVNEVMR
Mvan_5256|M.vanbaalenii_PYR-1 QEVGRLAEEIVGGYGNPADPETAELLDWLDKMTKRNLVITFGGGVNEVMR
Mflv_1510|M.gilvum_PYR-GCK QEVGRLAEELVGRYGNPADAHTSELLDWLDKMTKRNLVITFGGGVNEVMR
Mb3573c|M.bovis_AF2122/97 QRAGRLAEEIVGKYGNPAEPDTAELLRWLDAQTKRNLVITFGGGVNEVMR
Rv3543c|M.tuberculosis_H37Rv QRAGRLAEEIVGKYGNPAEPDTAELLRWLDAQTKRNLVITFGGGVNEVMR
MMAR_5030|M.marinum_M QRVGRLAEEIVGKYGNPAESDTAELLEWLDAQTKRNLVITFGGGVNEVMR
MUL_4104|M.ulcerans_Agy99 QRVGRLAEEIVGKYGNPAESDTAELLEWLDAQTKRNLVITFGGGVNEVMR
MAV_0618|M.avium_104 QYIGRLAEEVVGKYGNPADPETGELLRWLDSQTKRNLVITFGGGVNEVMR
TH_4261|M.thermoresistible__bu QKVGRLAEEIVGGYGDPADPETAELLDWLDKQTKRNLVITFGGGVNEVMR
MAB_0615|M.abscessus_ATCC_1997 QKLGRLSEGVVGRYGDPADANTAETLEWLDAQTKRHLVITFGGGVNEVMR
MLBr_00737|M.leprae_Br4923 MEVTTDAVQLFGGAGYTSDFP-------VERFMRDAKITQIYEGTNQIQR
: :.. * .:: :: : : : *.*:: *
MSMEG_5993|M.smegmatis_MC2_155 EMIAASGLKVPRVPR
Mvan_5256|M.vanbaalenii_PYR-1 EMIAASGLKVPRVPR
Mflv_1510|M.gilvum_PYR-GCK EMIAASGLKVPRVPR
Mb3573c|M.bovis_AF2122/97 EMIAASGLKVPRVPR
Rv3543c|M.tuberculosis_H37Rv EMIAASGLKVPRVPR
MMAR_5030|M.marinum_M EMIAAAGLKVPRVPR
MUL_4104|M.ulcerans_Agy99 EMIAAAGLKVPRVPR
MAV_0618|M.avium_104 EMIAAAGLKVPRVPR
TH_4261|M.thermoresistible__bu EMIAASGLKVPRVPR
MAB_0615|M.abscessus_ATCC_1997 EMVSTAGLGTPRVPR
MLBr_00737|M.leprae_Br4923 VVMSRALLR------
::: : *