For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSAAAEAWGNLQAIRVAVLVGFMPVLLYRIWRVARFPASVPALAATAFGASAWFWLLIYSDWVWPSLPPV VHAVSVAGWPPITIAACLQVFVVGIAGDASPARIRRALRTASVMAVLALIAVTVLVTRSRLPMSSDDVYV LAEAVFTEGDPAAVTAVVISSAYVIFVAVQLAWHGFRHIDRTPVGVGLGLMAVASIFQIVACVFGGIWQP LSRGQGVMGSAYGVWLDTWPGCIAEILMFTGFLWPPVVSRVQAHRDVRRLRPLHNALADLFPALFPPMES RIRLSDKVFEWGFHIQDGLTLLAQRRGDPLVAGRPAAEESSARAGVVAKWLVGQPVPGFSREWLHAPDGV SDESWMLAIADAYRERQEDAEFPASLSGMPSTLRR
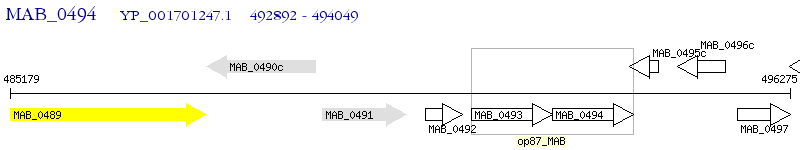
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0494 | - | - | 100% (385) | hypothetical protein MAB_0494 |
| M. abscessus ATCC 19977 | MAB_0493 | - | 6e-93 | 47.37% (361) | hypothetical protein MAB_0493 |
| M. abscessus ATCC 19977 | MAB_0114 | - | 3e-14 | 26.15% (325) | hypothetical protein MAB_0114 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1132 | - | 1e-10 | 25.00% (332) | hypothetical protein Mflv_1132 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6430 | - | 1e-12 | 25.90% (332) | hypothetical protein MSMEG_6430 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5674 | - | 1e-13 | 24.93% (349) | hypothetical protein Mvan_5674 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1132|M.gilvum_PYR-GCK --MIVWVIAGLLGLATGLRIGWALVNKQSLVSTAMILALG-SLGVVAALN
Mvan_5674|M.vanbaalenii_PYR-1 --MIVWVIAGLLGLATGLRIGWALVNKQSLVSTAMILALG-SLGLVAALN
MSMEG_6430|M.smegmatis_MC2_155 --MIIWVIAGLLGLATGLRIGWALVNKQSLVSSAMIVALG-SLGAVAALN
MAB_0494|M.abscessus_ATCC_1997 MSAAAEAWGNLQAIRVAVLVGFMPVLLYRIWRVARFPASVPALAATAFGA
. ..* .: ..: :*: * : * : * :*. .*
Mflv_1132|M.gilvum_PYR-GCK WEPLTLLIDTMLRWPNIS-----MGLSQVALIACAAGSAVMITTVASSRT
Mvan_5674|M.vanbaalenii_PYR-1 WQPLTLLIDTLLRWPNIS-----IGLSQVALVACAAGSCVMITTVSSSRA
MSMEG_6430|M.smegmatis_MC2_155 WQPLALLIDTLLRWPNLA-----VGLSQVALVACAAGSCVMITSASSERS
MAB_0494|M.abscessus_ATCC_1997 SAWFWLLIYSDWVWPSLPPVVHAVSVAGWPPITIAACLQVFVVGIAGDAS
: *** : **.:. :.:: . :: ** *::. :.. :
Mflv_1132|M.gilvum_PYR-GCK PATAKKLAFIQYGVAAVIAAVSLGIFLAAEQQPEMSPEEYLRRN----LS
Mvan_5674|M.vanbaalenii_PYR-1 PATARRIAVAQYGVAAVIAAVSLAVFLSAGQQPEMSPEEYLRRN----LG
MSMEG_6430|M.smegmatis_MC2_155 PVTTRRIAFAQYGIAAVVAVISLVMFFLAGRQPEMAPEEYLKHQ----LG
MAB_0494|M.abscessus_ATCC_1997 PARIRR-ALRTASVMAVLALIAVTVLVTRSRLPMSSDDVYVLAEAVFTEG
*. :: *. .: **:* ::: ::. : * : : *: : .
Mflv_1132|M.gilvum_PYR-GCK GGGNMLPWLLPLLYVLVALSLVAAAGLRHSNRTRRGRALFVFTIGIVLIV
Mvan_5674|M.vanbaalenii_PYR-1 SGGNALPWLLPLLYVLMALSLVVWAGLRHSNRTRRGRALFVFTVGIVLIV
MSMEG_6430|M.smegmatis_MC2_155 DNG--MSWLLPLLYVLLALTLVAWAGLRYSNRSRRGRALFVFTVGMVLIV
MAB_0494|M.abscessus_ATCC_1997 DPAAVTAVVISSAYVIFVAVQLAWHGFRHIDRTPVGVGLGLMAVASIFQI
. . . ::. **:.. :. *:*: :*: * .* ::::. :: :
Mflv_1132|M.gilvum_PYR-GCK LALAF-----FLLRAVGTSG-LVGVGAAATLLGCAMLIVAAGSLLPSLED
Mvan_5674|M.vanbaalenii_PYR-1 LASAF-----FLLRAAGTTG-PVGVGAAATLLGCAMLIVAAGSLLPSLED
MSMEG_6430|M.smegmatis_MC2_155 AASAF-----FLLRAVGTSD-TIGVGAAATLLGCAMVVVAAGSLLPSIED
MAB_0494|M.abscessus_ATCC_1997 VACVFGGIWQPLSRGQGVMGSAYGVWLDTWPGCIAEILMFTGFLWPPVVS
* .* * *. *. . ** : * ::: :* * *.: .
Mflv_1132|M.gilvum_PYR-GCK WVGARRELRTIAPLLEELGRRHPDVGIGVRPRGPLVFRVAERMSLISDAL
Mvan_5674|M.vanbaalenii_PYR-1 WFGAHRELRTIQPILDELGHRHPDVGIGVRPRGPLAFRVAERMSLISDAL
MSMEG_6430|M.smegmatis_MC2_155 WFGARQELRVIAPLLDELGKRQPDVGIGVRPRGPLVFRVAERMSLISDAL
MAB_0494|M.abscessus_ATCC_1997 RVQAHRDVRRLRPLHNALADLFPALFPPMESRIRLSDKVFEWGFHIQDGL
. *::::* : *: : *. * : :..* * :* * *.*.*
Mflv_1132|M.gilvum_PYR-GCK FLEATAADGALRPVDSAGAEVMDLDDLVERAELEAPDVAPSEQARAVAAW
Mvan_5674|M.vanbaalenii_PYR-1 FLEATAADGALRPVDSSGADVTDLEDLIAREDLEAPDVPPAEQARAVAEW
MSMEG_6430|M.smegmatis_MC2_155 YLEATAAESRRRKAARHGSGSAD------RPVLDVPEVTPQEQARAVAQW
MAB_0494|M.abscessus_ATCC_1997 TLLAQRRGDPLVAGRPAAE-------------------ESSARAGVVAKW
* * . . . :* .** *
Mflv_1132|M.gilvum_PYR-GCK IWDGR--------EKFPGLA--WLRQPARYSDREWILEIARQYRELS---
Mvan_5674|M.vanbaalenii_PYR-1 IFAGRGGAAGDGRAKFPGLA--WLRQPSTFSDREWILEIARQYRTLT---
MSMEG_6430|M.smegmatis_MC2_155 IHDTRDEDAP--ASAFPGLS--WLGQPESLSDREWILAIAQQYREIERAG
MAB_0494|M.abscessus_ATCC_1997 LVGQP----------VPGFSREWLHAPDGVSDESWMLAIADAYRERQ---
: .**:: ** * **..*:* ** **
Mflv_1132|M.gilvum_PYR-GCK -------TGFS------AGD------------------------------
Mvan_5674|M.vanbaalenii_PYR-1 -------ADAVHAVPERAGQGAVAD-------------------------
MSMEG_6430|M.smegmatis_MC2_155 TPSDSPERDLLTLHDIIAGRGPESSLRGVELFAAAQFLGLLDQVLLSLRR
MAB_0494|M.abscessus_ATCC_1997 --------EDAEFPASLSGMPSTLRR------------------------
:*
Mflv_1132|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5674|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6430|M.smegmatis_MC2_155 QSDGAGSNAADPFVTHVREPGEFLVELLVVLVVGEVRGLDTEEVGKSSHG
MAB_0494|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1132|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5674|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6430|M.smegmatis_MC2_155 CGGRVGAVPRTQLRKIRSRHGDALGLERSNHLGRRVGSAPGGINRVEQAV
MAB_0494|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1132|M.gilvum_PYR-GCK --------------------------------------------
Mvan_5674|M.vanbaalenii_PYR-1 --------------------------------------------
MSMEG_6430|M.smegmatis_MC2_155 QTGGERLAHRFDLHMIHERNEKAAYLLTIVPSVVTTTKCKPRKS
MAB_0494|M.abscessus_ATCC_1997 --------------------------------------------