For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGNGFRTLEENQKVEFEVGQSPKGPQATGVRAV
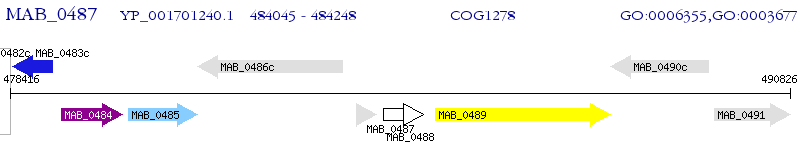
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0487 | - | - | 100% (67) | cold shock protein A (CspA) |
| M. abscessus ATCC 19977 | MAB_0872 | - | 7e-11 | 43.28% (67) | cold shock-like protein B CspB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3672c | cspA | 7e-34 | 92.54% (67) | cold shock protein A |
| M. gilvum PYR-GCK | Mflv_1392 | - | 3e-35 | 98.51% (67) | cold-shock DNA-binding domain-containing protein |
| M. tuberculosis H37Rv | Rv3648c | cspA | 7e-34 | 92.54% (67) | cold shock protein A |
| M. leprae Br4923 | MLBr_00198 | cspA | 9e-35 | 94.03% (67) | putative cold shock protein |
| M. marinum M | MMAR_5140 | cspA_1 | 3e-34 | 94.03% (67) | cold shock protein A CspA_1 |
| M. avium 104 | MAV_0516 | - | 8e-34 | 92.54% (67) | hypothetical protein MAV_0516 |
| M. smegmatis MC2 155 | MSMEG_6159 | - | 1e-34 | 95.52% (67) | hypothetical protein MSMEG_6159 |
| M. thermoresistible (build 8) | TH_0855 | - | 2e-31 | 93.55% (62) | PROBABLE COLD SHOCK PROTEIN A CSPA |
| M. ulcerans Agy99 | MUL_4224 | cspA_1 | 2e-34 | 94.03% (67) | cold shock protein A CspA_1 |
| M. vanbaalenii PYR-1 | Mvan_5398 | - | 4e-35 | 97.01% (67) | cold-shock DNA-binding domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0487|M.abscessus_ATCC_1997 MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGNGFRTLEENQKVEF
Mflv_1392|M.gilvum_PYR-GCK MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGSGFRTLEENQKVEF
MSMEG_6159|M.smegmatis_MC2_155 MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGSGFRTLEENQKVEF
TH_0855|M.thermoresistible__bu -----VKWFNAEKGFGFIAPEDGSADVFVHYTEIQGSGFRTLEENQRVEF
Mvan_5398|M.vanbaalenii_PYR-1 MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGSGFRTLEENQKVEF
Mb3672c|M.bovis_AF2122/97 MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGTGFRTLEENQKVEF
Rv3648c|M.tuberculosis_H37Rv MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGTGFRTLEENQKVEF
MAV_0516|M.avium_104 MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGTGFRTLEENQKVEF
MLBr_00198|M.leprae_Br4923 MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGSGFRTLEENQKVEF
MMAR_5140|M.marinum_M MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGSGFRTLEENQKVEF
MUL_4224|M.ulcerans_Agy99 MPQGTVKWFNAEKGFGFIAPEDGSADVFVHYTEIQGSGFRTLEENQKVEF
*******************************.*********:***
MAB_0487|M.abscessus_ATCC_1997 EVGQSPKGPQATGVRAV
Mflv_1392|M.gilvum_PYR-GCK EVGQSPKGPQATGVRAV
MSMEG_6159|M.smegmatis_MC2_155 EVGQSPKGPQATGVRTI
TH_0855|M.thermoresistible__bu EVGQSPKGPQATGVRSI
Mvan_5398|M.vanbaalenii_PYR-1 EVGQSPKGPQATGVRAI
Mb3672c|M.bovis_AF2122/97 EIGHSPKGPQATGVRSL
Rv3648c|M.tuberculosis_H37Rv EIGHSPKGPQATGVRSL
MAV_0516|M.avium_104 EIGHSPKGPQATGVRSL
MLBr_00198|M.leprae_Br4923 EIGHSPKGPQATGVRSV
MMAR_5140|M.marinum_M EIGHSPKGPQATGVRSV
MUL_4224|M.ulcerans_Agy99 EIGHSPKGPQATGVRSV
*:*:***********::