For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTHSARLAELGIELPAVAAPLAAYQPAVRSGNHVYTSGQLPMVDGNLATAGKVGAEVSPEDAKALARIC ALNAVAAVDALVGINNVVKVVKVVGFVASAPGFTGQPGVVNGASELLGEIFGEAGVHARSAVGVAELPIN APVEVEIVVEVREEVSA
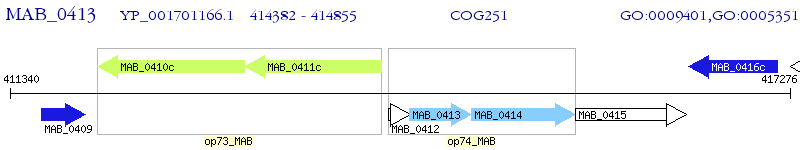
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0413 | - | - | 100% (157) | putative endoribonuclease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3702c | - | 9e-56 | 69.86% (146) | hypothetical protein Mb3702c |
| M. gilvum PYR-GCK | Mflv_1369 | - | 1e-59 | 80.14% (146) | endoribonuclease L-PSP |
| M. tuberculosis H37Rv | Rv3678c | - | 9e-56 | 69.86% (146) | hypothetical protein Rv3678c |
| M. leprae Br4923 | MLBr_02304 | - | 2e-53 | 69.18% (146) | hypothetical protein MLBr_02304 |
| M. marinum M | MMAR_5166 | - | 5e-57 | 72.11% (147) | hypothetical protein MMAR_5166 |
| M. avium 104 | MAV_0451 | - | 2e-59 | 75.34% (146) | translation initiation inhibitor |
| M. smegmatis MC2 155 | MSMEG_6191 | - | 1e-62 | 81.63% (147) | translation initiation inhibitor |
| M. thermoresistible (build 8) | TH_2127 | - | 1e-63 | 77.63% (152) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4252 | - | 4e-57 | 72.11% (147) | hypothetical protein MUL_4252 |
| M. vanbaalenii PYR-1 | Mvan_5437 | - | 6e-61 | 78.15% (151) | endoribonuclease L-PSP |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1369|M.gilvum_PYR-GCK -------------------MSSG--WQARLDELGIELPDVVAPLAAYVPA
Mvan_5437|M.vanbaalenii_PYR-1 --------------------MSG--WSERLDELGIALPEVVAPLAAYVPA
MSMEG_6191|M.smegmatis_MC2_155 --------------------MSGASWSARLAELGIELPGVVPPVAAYVPA
MAB_0413|M.abscessus_ATCC_1997 --------------------MST--HSARLAELGIELPAVAAPLAAYQPA
TH_2127|M.thermoresistible__bu --------------------VST--WSNRLTELGISLPAVAAPVAAYVPA
Mb3702c|M.bovis_AF2122/97 -----------------------MSAKARLGQLGVTLPQVAAPLAAYVPA
Rv3678c|M.tuberculosis_H37Rv -----------------------MSAKARLGQLGVTLPQVAAPLAAYVPA
MMAR_5166|M.marinum_M -----------------------MNATSRLAELGITLPQVAAPLAAYVPA
MUL_4252|M.ulcerans_Agy99 -----------------------MNATSRLAELGITLPQVAAPLAAYVPA
MLBr_02304|M.leprae_Br4923 MGVSGGAARPHGRAACCILEASEMSVLARLGQLGIELPQAAPPLASYVSA
MAV_0451|M.avium_104 -------------------MSAAPSWKARLAELGLTLPEVVAPLASYVPA
** :**: ** ...*:*:* .*
Mflv_1369|M.gilvum_PYR-GCK VRTGNLVYTSGQLPIRDGELLAAGKVGAEVSAEDAKPLARQCALNALAAV
Mvan_5437|M.vanbaalenii_PYR-1 VRTGNLVYTAGQLPISNGELAATGKVGAEVSAEDAKHLARRCALNALAAV
MSMEG_6191|M.smegmatis_MC2_155 VHTGNHVYTSGQLPIRGGELIKAGKVGAEVSAEEAKELARVCGLNALAAV
MAB_0413|M.abscessus_ATCC_1997 VRSGNHVYTSGQLPMVDGNLATAGKVGAEVSPEDAKALARICALNAVAAV
TH_2127|M.thermoresistible__bu VRTGNLVYTSGQLPMVDGNLAATGKVGGEVTPEQAKALARVCALNALAAV
Mb3702c|M.bovis_AF2122/97 VRTGNLVYTAGQLPLEAGKLVRTGKLGADVNPEEGKTLARICALNALAAV
Rv3678c|M.tuberculosis_H37Rv VRTGNLVYTAGQLPLEAGKLVRTGKLGADVNPEEGKTLARICALNALAAV
MMAR_5166|M.marinum_M VRTGNLIYTAGQLPLEAGKLLGTGKVGANVTPEEGKALARVCALNALAAV
MUL_4252|M.ulcerans_Agy99 VRTGNLIYTAGQLPLEAGKLLGTGKVGANVTPEEGKALARVCALNALAAV
MLBr_02304|M.leprae_Br4923 VRTGNLVYTSGQLPIEDGRLPQTGKVGADVSLGEGKALARICALNALAAV
MAV_0451|M.avium_104 VRTGNLVYTAGQLPMQDGKLATTGKVGAHITPDEGRALARICALNALAAV
*::** :**:****: *.* :**:*..:. :.: *** *.***:***
Mflv_1369|M.gilvum_PYR-GCK HALTGIESVVKVVKVVGFVASAPGFDGQPGVINGASELFGEIFGEAGAHA
Mvan_5437|M.vanbaalenii_PYR-1 DALVGIDSVVKVVKVVGFVASAPGFDGQPGVINGASELFGEVFGEAGAHA
MSMEG_6191|M.smegmatis_MC2_155 HALVGIDSVVKVVKVVGFVASAPGFGGQPGVVNGASELFGEIFGDAGAHA
MAB_0413|M.abscessus_ATCC_1997 DALVGINNVVKVVKVVGFVASAPGFTGQPGVVNGASELLGEIFGEAGVHA
TH_2127|M.thermoresistible__bu HDLAGIDAVTRIVKVVGFVASAPGFTGQPGVVNGASELFGEIFGDAGAHA
Mb3702c|M.bovis_AF2122/97 DSLVDLDAVTRVVKVVGFVASAPGFHGQPSVINGASDLLAEVFGDSGAHA
Rv3678c|M.tuberculosis_H37Rv DSLVDLDAVTRVVKVVGFVASAPGFHGQPSVINGASDLLAEVFGDSGAHA
MMAR_5166|M.marinum_M NSLVDIDSVTRVVKVVGFVASAPGFHGQPNVINGASDLLAEVFGDKGVHA
MUL_4252|M.ulcerans_Agy99 NSLVDIDSVTRVVKVVGFVASAPGFHGQPNVINGASDLLAEVFGDKGVHA
MLBr_02304|M.leprae_Br4923 DSLVGLDSVTGVVKVVGFVASSSGFNGQPSVVNGASDLFAEVFGDRGRHA
MAV_0451|M.avium_104 DALVGIDAVTRVVKVVGFVASAPGFNGQPSVVNGASDLLAEVFGEHGAHA
. *..:: *. :*********:.** ***.*:****:*:.*:**: * **
Mflv_1369|M.gilvum_PYR-GCK RSAVGVSELPRNTPVEVEAIFEVA-----
Mvan_5437|M.vanbaalenii_PYR-1 RSAVGVSELPRNAPVEVEVIVEVS-----
MSMEG_6191|M.smegmatis_MC2_155 RSAVGVAELPINAPVEVEIIVEVTPGLA-
MAB_0413|M.abscessus_ATCC_1997 RSAVGVAELPINAPVEVEIVVEVREEVSA
TH_2127|M.thermoresistible__bu RSAVGVSELPLDAPVEVELIAEIR-----
Mb3702c|M.bovis_AF2122/97 RSAVGVSELPLDAPVEVELIVEVG-----
Rv3678c|M.tuberculosis_H37Rv RSAVGVSELPLDAPVEVELIVEVG-----
MMAR_5166|M.marinum_M RSAVGVAELPLDAPVEVELIVEVG-----
MUL_4252|M.ulcerans_Agy99 RSAVGVAELPLDAPVEVELIVEVG-----
MLBr_02304|M.leprae_Br4923 RSAVGVAELPLGAPVEIELIVELG-----
MAV_0451|M.avium_104 RSAVGVGELPLDAPVEVELIVEVGARP--
******.*** .:***:* : *: