For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNTSPDSPSDAAAGPPLTTWPARVANARLHFVTGKGGTGKSTIAAALALALAAGGRKVLLVEVENRQGIA QLFDVPPLAPKELKIATAEGGGVVNALAIEIETAFMEYLDMFYNLGLAGRAMKRIGAIEFATTIAPGLRD VILTGKIKECIVRTDKSGRHVYDAVVVDAPPTGRIARFLDVTSALSDIAKGGPIHSQSEGVRRLLHSDQT AIHLVTLLEALPMQETAEAIAELSEMGLPVGSVIVNRTVVAHLSPADLDKAAEGVIDADAVRKALDENNV TLSDADFAGLLTETIEHATRMRARAESEELLGELHIPRLELPTLADGVDLGSLYELADHLAEQGVR*
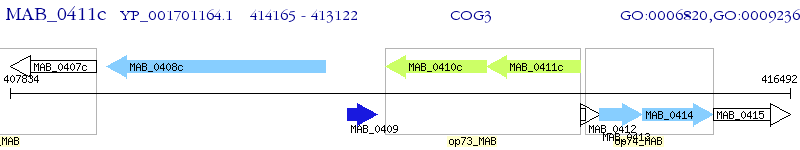
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0411c | - | - | 100% (347) | putative anion transporter ATPase |
| M. abscessus ATCC 19977 | MAB_1982 | - | 2e-08 | 22.66% (278) | putative arsenite-transporting ATPase |
| M. abscessus ATCC 19977 | MAB_0410c | - | 7e-07 | 24.68% (158) | anion transporter ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3704 | - | 1e-139 | 74.93% (335) | anion transporter ATPase |
| M. gilvum PYR-GCK | Mflv_1368 | - | 1e-142 | 75.00% (340) | anion-transporting ATPase |
| M. tuberculosis H37Rv | Rv3679 | - | 1e-139 | 74.93% (335) | anion transporter ATPase |
| M. leprae Br4923 | MLBr_02305 | - | 1e-133 | 70.93% (344) | putative anion transporter protein |
| M. marinum M | MMAR_5168 | - | 1e-140 | 74.26% (338) | anion transporter ATPase |
| M. avium 104 | MAV_0449 | - | 1e-137 | 76.78% (323) | anion-transporting ATPase |
| M. smegmatis MC2 155 | MSMEG_6193 | - | 1e-138 | 75.60% (332) | anion-transporting ATPase |
| M. thermoresistible (build 8) | TH_2125 | - | 1e-138 | 72.51% (342) | PROBABLE ANION TRANSPORTER ATPASE |
| M. ulcerans Agy99 | MUL_4254 | - | 1e-139 | 73.96% (338) | anion transporter ATPase |
| M. vanbaalenii PYR-1 | Mvan_5439 | - | 1e-139 | 74.71% (344) | anion-transporting ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3704|M.bovis_AF2122/97 MVATTSS-------GGSSVGWPSRLSGVRLHLVTGKGGTGKSTIAAALAL
Rv3679|M.tuberculosis_H37Rv MVATTSS-------GGSSVGWPSRLSGVRLHLVTGKGGTGKSTIAAALAL
MMAR_5168|M.marinum_M MVANTSS-------GSGSVGWPSRLSKARLHFVTGKGGTGKSTIAAALAL
MUL_4254|M.ulcerans_Agy99 MVANTSS-------GSGSVGWTSRLSKARLHFVTGKGGTGKSTIAAALAL
MLBr_02305|M.leprae_Br4923 MVTPTSG-------KDSAIGWPTRLSKARLHLVTGKGGTGKSTIAAALAL
MAV_0449|M.avium_104 ------------------------MTKARLHFVTGKGGTGKSTIAAALAL
Mflv_1368|M.gilvum_PYR-GCK MATTYTG----TTDGPKPVGWPSRLTKARLHFVSGKGGTGKSTIAAALAL
Mvan_5439|M.vanbaalenii_PYR-1 MATTSSASS--VPDGTKQLGWPSRLTKARLHFVSGKGGTGKSTIAAALAL
TH_2125|M.thermoresistible__bu --VVSSS-----PSGNKPVGWPSRLTRARLHFVTGKGGTGKSTIAAALAL
MSMEG_6193|M.smegmatis_MC2_155 MATTESG--------AKHVGWPSRLTKARLHFVSGKGGTGKSTIAAALAL
MAB_0411c|M.abscessus_ATCC_199 MVNTSPDSPSDAAAGPPLTTWPARVANARLHFVTGKGGTGKSTIAAALAL
:: .***:*:****************
Mb3704|M.bovis_AF2122/97 TLAAGGRKVLLVEVEGRQGIAQLFDVPPLPYQELKIATAERGGQVNALAI
Rv3679|M.tuberculosis_H37Rv TLAAGGRKVLLVEVEGRQGIAQLFDVPPLPYQELKIATAERGGQVNALAI
MMAR_5168|M.marinum_M TLAAGGRKVLLVEVEGRQGIAQLFDVPPLPYQEVKIATAERGGQVNALAI
MUL_4254|M.ulcerans_Agy99 TLAAGGRKVLLVEVEGRQGIAQLFDVPPLPYQEVKIATAERGGQVNALAI
MLBr_02305|M.leprae_Br4923 TLAAGGRKVLLIEVEGRQGIAQLFDVPPLPYQELKIATAAGGGQVNALAI
MAV_0449|M.avium_104 TLAAGGRKVLLVEVEGRQGIAQLFDVPPLPYQEVKIATAERGGQVNALAI
Mflv_1368|M.gilvum_PYR-GCK TLAAGGRKVLLVEVEGRQGIAQLFDVPPLPYEEVKIATADGGGQVNALAI
Mvan_5439|M.vanbaalenii_PYR-1 ALAAGGRKVLLVEVEGRQGIAQLFDVPPLPYEEVKIATAEGGGQVNALAI
TH_2125|M.thermoresistible__bu CLAAGGRKVLLVEVEGRQGIAQLFDVPPLPYEEVKIATAEGGGVVNALAI
MSMEG_6193|M.smegmatis_MC2_155 ALAAHGRRVLLVEVEERQGIAQLFDVPPLPYEEVKIATAEGGGQVNALAI
MAB_0411c|M.abscessus_ATCC_199 ALAAGGRKVLLVEVENRQGIAQLFDVPPLAPKELKIATAEGGGVVNALAI
*** **:***:*** *************. :*:***** ** ******
Mb3704|M.bovis_AF2122/97 DIEAAFLEYLDMFYNLGIAGRAMRRIGAVEFATTIAPGLRDVLLTGKIKE
Rv3679|M.tuberculosis_H37Rv DIEAAFLEYLDMFYNLGIAGRAMRRIGAVEFATTIAPGLRDVLLTGKIKE
MMAR_5168|M.marinum_M DIEAAFLEYLEMFYNLGIAGRAMRRIGAIEFATTIAPGLRDVLLTGKIKE
MUL_4254|M.ulcerans_Agy99 DIEAAFLEYLEMFYNLGIAGRAMRRIGAIEFATTIAPGLRDVLLTGKIKE
MLBr_02305|M.leprae_Br4923 DIEAAFLEYLDMFYNLGLAGRAMRRIGAIEFATTIAPGLRDVLLTGKIKE
MAV_0449|M.avium_104 DIEAAFLEYLDMFYNLGIAGRAMRRIGAIEFATTIAPGLRDVLLTGKIKE
Mflv_1368|M.gilvum_PYR-GCK DTEAAFLEYLDMFYNLGLAGRAMRRIGAVEFATTIAPGLRDVLLTGKIKE
Mvan_5439|M.vanbaalenii_PYR-1 DTEAAFLEYLDMFYNLGLAGRAMRRIGAVEFATTIAPGLRDVLLTGKIKE
TH_2125|M.thermoresistible__bu DTEAAFLEYLDMFYNLGLAGRAMRRIGAVEFATTIAPGLRDVLLTGKIRE
MSMEG_6193|M.smegmatis_MC2_155 DIEAAFLEYLDMFYNLGLAGRAMRRIGAIEFATTIAPGLRDVLLTGKIKE
MAB_0411c|M.abscessus_ATCC_199 EIETAFMEYLDMFYNLGLAGRAMKRIGAIEFATTIAPGLRDVILTGKIKE
: *:**:***:******:*****:****:*************:*****:*
Mb3704|M.bovis_AF2122/97 TVVRLDK-------NKLPVYDAIVVDAPPTGRIARFLDVTKAVSDLAKGG
Rv3679|M.tuberculosis_H37Rv TVVRLDK-------NKLPVYDAIVVDAPPTGRIARFLDVTKAVSDLAKGG
MMAR_5168|M.marinum_M TVVRVDK-------NRLPVYDAIVVDSPPTGRIARFLDVTKAVSDLAKGG
MUL_4254|M.ulcerans_Agy99 TVVRVDK-------NRLPVYDAIVVDSPPTGRIARFLDVTKAVSDLAKGG
MLBr_02305|M.leprae_Br4923 SVVRINKG------AKNPVYDAIVVDAPPTGRITRFLDVTKAVSNLAKDG
MAV_0449|M.avium_104 TVIRVDK-------NRLPVYDAIVVDAPPTGRIARFLDVTKAVSDLAKGG
Mflv_1368|M.gilvum_PYR-GCK IVTRNDKAGKADKSGKGTAYDAVVVDSPPTGRIARFLDVTKAVSDLAKGG
Mvan_5439|M.vanbaalenii_PYR-1 IVTRNEKD---QPRGKGHVYDAVVVDSPPTGRIARFLDVTKAVSDLAKGG
TH_2125|M.thermoresistible__bu IVTRNDKP---DKHAKQQIYDAVVVDSPPTGRISRFLDVTKAVSDLAKGG
MSMEG_6193|M.smegmatis_MC2_155 IVVRPKTMR--DEKHKRSGYDAVVVDSPPTGRISRFLDVTKAVSDLAKGG
MAB_0411c|M.abscessus_ATCC_199 CIVRTDK-------SGRHVYDAVVVDAPPTGRIARFLDVTSALSDIAKGG
: * .. ***:***:******:******.*:*::**.*
Mb3704|M.bovis_AF2122/97 PVHAQSEGVVKLLHSNQTAIHLVTLLEALPVQETLEAIEELAQMELPIGS
Rv3679|M.tuberculosis_H37Rv PVHAQSEGVVKLLHSNQTAIHLVTLLEALPVQETLEAIEELAQMELPIGS
MMAR_5168|M.marinum_M PVHSQAEGVVNLLHSDQTAIHLVTLLEALPVQETLEAIEELAEMELPIGS
MUL_4254|M.ulcerans_Agy99 PVHSQAEGVVNLLHSDQTAIHLVTLLEALPVQETLEAIEELAEMELPIGS
MLBr_02305|M.leprae_Br4923 PVHSQSQGVVELLHSDQTAIHLVTLLEALPVQETLEAIQELADMELPIGS
MAV_0449|M.avium_104 PVHSQADGVVRLLHSEQTAIHLVTLLEALPVQETLEAIEELAEMQLPIGS
Mflv_1368|M.gilvum_PYR-GCK PVHSQAEGVVKLLHSDLTAIHLVTLLEALPIQETLEAIDELREMGLPIGS
Mvan_5439|M.vanbaalenii_PYR-1 PVHSQADGVVKLLHSDLTAIHLVTLLEALPIQETLEAIDELREMDLPIGS
TH_2125|M.thermoresistible__bu PVHAQAESVVNVLHSDLTAIHLVTLLEALPMQETIEAIEELRELDLPIGS
MSMEG_6193|M.smegmatis_MC2_155 PVHSQAEGVVKLLHSEQTAIHLVTLLEALPIQETLEAIDELKELNLPIGS
MAB_0411c|M.abscessus_ATCC_199 PIHSQSEGVRRLLHSDQTAIHLVTLLEALPMQETAEAIAELSEMGLPVGS
*:*:*::.* .:***: *************:*** *** ** :: **:**
Mb3704|M.bovis_AF2122/97 VIVNRNIPAHLEPQDLAKAAEGEVDADSVRAGLLTAGVKLPDADFAGLLT
Rv3679|M.tuberculosis_H37Rv VIVNRNIPAHLEPQDLAKAAEGEVDADSVRAGLLTAGVKLPDADFAGLLT
MMAR_5168|M.marinum_M VIVNRNIPAYLDPGDLAKAAEGDVDADSVRSGLAMAGIELADADFAGLLT
MUL_4254|M.ulcerans_Agy99 VIVNRNIPAYLDPGDLAKAAEGDVDADSVRSGLAMAGIELADADFAGLLT
MLBr_02305|M.leprae_Br4923 VIVNRNIPCYLGHHDLAKVAEGDIDADTVRKGLAMAGIKLNDADFAGLLT
MAV_0449|M.avium_104 VIVNRNIPAYLQPADLAKAAEGDIDADAVRAGLQKAGITLDDKDFAGLLT
Mflv_1368|M.gilvum_PYR-GCK VIVNRNISSYLAPDDLAKAADGVVDADAVRAGLESAGITLSDNDFAGLLT
Mvan_5439|M.vanbaalenii_PYR-1 VIVNRNIPAYLAPDDLAKAAEGVIDADAMRAGLTTAGITLSDDDFAGLLT
TH_2125|M.thermoresistible__bu VIVNRNIPAYLAPGDLAKAAEGDIDADTVRTDLAKTGITLSDKDFAGLLT
MSMEG_6193|M.smegmatis_MC2_155 VIVNRNIPAYLSPDDLAKAAEGDLDADTIRADLSKAGITLSDENFAGLLT
MAB_0411c|M.abscessus_ATCC_199 VIVNRTVVAHLSPADLDKAAEGVIDADAVRKALDENNVTLSDADFAGLLT
*****.: .:* ** *.*:* :***::* * .: * * :******
Mb3704|M.bovis_AF2122/97 ETIQHATRITARAEIAQQLDALQVPRLELPTVSDGVDLGSLYELSESLAQ
Rv3679|M.tuberculosis_H37Rv ETIQHATRITARAEIAQQLDALQVPRLELPTVSDGVDLGSLYELSESLAQ
MMAR_5168|M.marinum_M ETIEHATRITARAETAQQLDALQVARLELPTISDGVDLGSLYELSESLAQ
MUL_4254|M.ulcerans_Agy99 ETIEHATRITARAETAQQLDALQVARLELPTISDGVDLGSLYELSESLAQ
MLBr_02305|M.leprae_Br4923 ETIQYAMVITTRAEAAQQLDTLQVPRLDLPAISDGVDLGNLYELSESLAQ
MAV_0449|M.avium_104 ETIEHATVIATRAEIAQQLDALHVARLELPAISDGVDLGSLYELSESLAQ
Mflv_1368|M.gilvum_PYR-GCK ETIQHATRIRARTESAEQLAALDVPRLDLPTLPDGVDLGSLYELAEELAH
Mvan_5439|M.vanbaalenii_PYR-1 ETIQHATRIAARNESAAQLVELDVSRLDLPTLPDGVDLGSLYELAEELAH
TH_2125|M.thermoresistible__bu QAIQHATRIRARAESATQLDRLDVPRLQLPALPEGIDLGSLYELAEALAH
MSMEG_6193|M.smegmatis_MC2_155 EAIQHATRIKARKESAELLDEIDIPRLDLPALADGVDLGSLYELAEELAA
MAB_0411c|M.abscessus_ATCC_199 ETIEHATRMRARAESEELLGELHIPRLELPTLADGVDLGSLYELADHLAE
::*::* : :* * * :.:.**:**::.:*:***.****:: **
Mb3704|M.bovis_AF2122/97 QGVR
Rv3679|M.tuberculosis_H37Rv QGVR
MMAR_5168|M.marinum_M QGVR
MUL_4254|M.ulcerans_Agy99 QGVR
MLBr_02305|M.leprae_Br4923 QGVR
MAV_0449|M.avium_104 QGVR
Mflv_1368|M.gilvum_PYR-GCK QGVR
Mvan_5439|M.vanbaalenii_PYR-1 QGVR
TH_2125|M.thermoresistible__bu QGVR
MSMEG_6193|M.smegmatis_MC2_155 QGVR
MAB_0411c|M.abscessus_ATCC_199 QGVR
****