For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDVSADQEPLPLTTQAARFVITGGLSAIVDFGLYVLFLHVGLQVNVAKSISFVAGTTTAYLINRRWTFQA EPSRARFIAVMALYGLTYAVQVGINYLFYMQFEDKPWRVPVAFVIAQGTATVINFIVQRAVIFRIR*
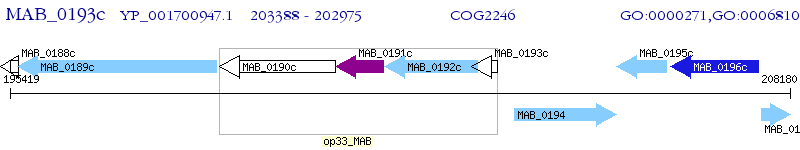
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0193c | - | - | 100% (137) | hypothetical protein MAB_0193c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3818 | - | 2e-45 | 72.50% (120) | hypothetical protein Mb3818 |
| M. gilvum PYR-GCK | Mflv_1179 | - | 4e-57 | 84.80% (125) | GtrA family protein |
| M. tuberculosis H37Rv | Rv3789 | - | 3e-45 | 72.50% (120) | integral membrane protein |
| M. leprae Br4923 | MLBr_00110 | - | 2e-38 | 62.81% (121) | hypothetical protein MLBr_00110 |
| M. marinum M | MMAR_5351 | - | 2e-42 | 66.67% (129) | hypothetical protein MMAR_5351 |
| M. avium 104 | MAV_0233 | - | 1e-46 | 71.09% (128) | membrane protein |
| M. smegmatis MC2 155 | MSMEG_6372 | - | 5e-56 | 81.89% (127) | membrane protein |
| M. thermoresistible (build 8) | TH_1999 | - | 4e-53 | 71.53% (137) | POSSIBLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4968 | - | 1e-41 | 65.12% (129) | hypothetical protein MUL_4968 |
| M. vanbaalenii PYR-1 | Mvan_5626 | - | 2e-57 | 82.68% (127) | GtrA family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1179|M.gilvum_PYR-GCK ------------MSP--QLSLTTQIWRFLITGGLSAIVDFGLYVALL-AA
Mvan_5626|M.vanbaalenii_PYR-1 ------------MSP--QLSLTTQIWRFVVTGGLSAVVDFGLYVALL-AA
MSMEG_6372|M.smegmatis_MC2_155 --------MAETTAP--RLSLGTQAFRFIVTGGLSAIVDFGLYVLLL-AA
TH_1999|M.thermoresistible__bu --------VAETAAPTTSLSLKTQVWRFLVTGGFSAVVDFGLYVLLL-VA
MAB_0193c|M.abscessus_ATCC_199 --------MSDVSADQEPLPLTTQAARFVITGGLSAIVDFGLYVLFL-HV
Mb3818|M.bovis_AF2122/97 -------------------------MRFVVTGGLAGIVDFGLYVVLYKVA
Rv3789|M.tuberculosis_H37Rv -------------------------MRFVVTGGLAGIVDFGLYVVLYKVA
MAV_0233|M.avium_104 ------------MVASPNLKLTTQVMRFVVTGGLAGIVDFGLYATLYKVV
MMAR_5351|M.marinum_M MIPPEPAAAPAEEHQRQHLHLTTQMIRFIVTGGFAAIVDFSVYVVLYKVV
MUL_4968|M.ulcerans_Agy99 MIPPEPAAAPTEEHQRQHLHLTTQMIRFIVTGGFAAIVDFSVYVVLYKVV
MLBr_00110|M.leprae_Br4923 ------------------------MLRFIVTGSLATAVDFSVYVTLYRGG
**::**.:: ***.:*. :
Mflv_1179|M.gilvum_PYR-GCK GLHVNIAKTISFIAGTTTAYLINRRWTFQAPPSKARFIAVCVLYGLTYAV
Mvan_5626|M.vanbaalenii_PYR-1 GLHVNIAKTLSFVAGTTTAYLINRRWTFQAAPSKARFIAVVALYAVTYAV
MSMEG_6372|M.smegmatis_MC2_155 GLHVNVAKTLSFVAGTTTAYLINRRWTFQAPPSKARFIAVCVLYAVTYAV
TH_1999|M.thermoresistible__bu GLHVNIAKTIGFIAGTTTAYLINRRWTFQAPPSTARFIAVCALYATTYAV
MAB_0193c|M.abscessus_ATCC_199 GLQVNVAKSISFVAGTTTAYLINRRWTFQAEPSRARFIAVMALYGLTYAV
Mb3818|M.bovis_AF2122/97 GLQVDLSKAISFIVGTITAYLINRRWTFQAEPSTARFVAVMLLYGITFAV
Rv3789|M.tuberculosis_H37Rv GLQVDLSKAISFIVGTITAYLINRRWTFQAEPSTARFVAVMLLYGITFAV
MAV_0233|M.avium_104 GVQVDVAKAISFIVGTITAYLINRRWTFQAAPSTARFVAVMALYAVTFAV
MMAR_5351|M.marinum_M GLQVDLSKFISVVIGTITAYLINRRWTFQAAPSGARFAAVMALYGVTFAV
MUL_4968|M.ulcerans_Agy99 GLQVDLSKFISVVIGTITAYLINRRWTFQAAPCGARFAAVMALYGVTFAV
MLBr_00110|M.leprae_Br4923 GLQVDLAKFTSVVIGTITSYMINRRWTFQMSPSTTRFAAVMALYGITFAV
*::*:::* ..: ** *:*:******** *. :** ** **. *:**
Mflv_1179|M.gilvum_PYR-GCK QVGINYVFYMEFDDKP-WRVPVAFVIAQGTATVINFIVQRAVIFRWAGA-
Mvan_5626|M.vanbaalenii_PYR-1 QVGINYVFYMEFEDQP-WRVPVAFVIAQGTATVINFIVQRAVIFRLR---
MSMEG_6372|M.smegmatis_MC2_155 QVGINYVFYMAWDEKP-WRVPVAFVIAQGTATVINFIVQRAVIFRLR---
TH_1999|M.thermoresistible__bu QVGINYLLYMAWEEQS-WRVPVAFVVAQGTATVINFVVQRTVIFRLRQPA
MAB_0193c|M.abscessus_ATCC_199 QVGINYLFYMQFEDKP-WRVPVAFVIAQGTATVINFIVQRAVIFRIR---
Mb3818|M.bovis_AF2122/97 QVGLNHLCLALLHYRA-WAIPVAFVIAQGTATVINFIVQRAVIFRIR---
Rv3789|M.tuberculosis_H37Rv QVGLNHLCLALLHYRA-WAIPVAFVIAQGTATVINFIVQRAVIFRIR---
MAV_0233|M.avium_104 QVGLNHLCLAFLHYRG-WAIPVAFVIAQGTATVINFVVQRAVIFRIR---
MMAR_5351|M.marinum_M QVGINHLCLALMHYRG-WAIPVAFVIAQAVATVINFVVQRAVIFRRH---
MUL_4968|M.ulcerans_Agy99 QVGINHLCLALMHYRG-WAIPVAFVIAQAVSTVINFVVQRAVIFRRH---
MLBr_00110|M.leprae_Br4923 QMGLNHLCLFLFHYQEPWAIPIAFVIAQGLATVINFIVQRVVIFRIR---
*:*:*:: . : * :*:***:**. :*****:***.****
Mflv_1179|M.gilvum_PYR-GCK -------
Mvan_5626|M.vanbaalenii_PYR-1 -------
MSMEG_6372|M.smegmatis_MC2_155 -------
TH_1999|M.thermoresistible__bu AGRRSNR
MAB_0193c|M.abscessus_ATCC_199 -------
Mb3818|M.bovis_AF2122/97 -------
Rv3789|M.tuberculosis_H37Rv -------
MAV_0233|M.avium_104 -------
MMAR_5351|M.marinum_M -------
MUL_4968|M.ulcerans_Agy99 -------
MLBr_00110|M.leprae_Br4923 -------