For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATTDLHARTSSPFLELLHDQIRNEFTASQQYIAIAVYFDDADLPQLAKRFYAQAVEERNHAMMIIRYLID KNVAITIPGVDAVVTEFADARAPIALALEQEKRVTDQVTELARAARSAGDYYGEQFMQWFLKEQVEEVAL MDTLLTVAERADHDLFDLENFVARELARPVSPDPTAPTAAGGAL*
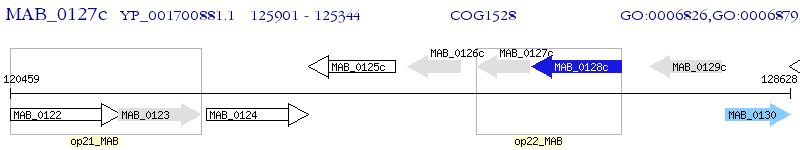
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0127c | - | - | 100% (185) | bacterioferritin BfrB |
| M. abscessus ATCC 19977 | MAB_0126c | - | 1e-65 | 73.84% (172) | bacterioferritin BfrB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3871 | bfrB | 3e-55 | 61.85% (173) | bacterioferritin BfrB |
| M. gilvum PYR-GCK | Mflv_1141 | - | 4e-56 | 62.21% (172) | Ferritin, Dps family protein |
| M. tuberculosis H37Rv | Rv3841 | bfrB | 3e-55 | 61.85% (173) | bacterioferritin BfrB |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5393 | bfrB | 2e-54 | 60.47% (172) | bacterioferritin BfrB |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6422 | - | 3e-59 | 63.95% (172) | ferritin family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_5014 | bfrB | 1e-54 | 60.47% (172) | bacterioferritin BfrB |
| M. vanbaalenii PYR-1 | Mvan_5666 | - | 6e-56 | 62.79% (172) | Ferritin, Dps family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3871|M.bovis_AF2122/97 ---------------MTEYEGPKTKFHALMQEQIHNEFTAAQQYVAIAVY
Rv3841|M.tuberculosis_H37Rv ---------------MTEYEGPKTKFHALMQEQIHNEFTAAQQYVAIAVY
MMAR_5393|M.marinum_M MPSLQVAFYLHYSGGMTDYDGPKTKFHALVQEQIFNEFTAAQQYVAIAVY
MUL_5014|M.ulcerans_Agy99 ---------------MTDYDGPKTKFHALVQEQIFNEFTAAQQYVAIAVY
MSMEG_6422|M.smegmatis_MC2_155 ---------------MTNSGALDTKFHALIQDQIRSEFTASQQYIAIAVF
Mflv_1141|M.gilvum_PYR-GCK MTATPFSTYS---------DSLDTKFHSLLNDQVRSELGASQQYLAVAVY
Mvan_5666|M.vanbaalenii_PYR-1 MNMQPDLVYLGEMTSPGALDAVDTKFHSLLNEQVRSELGASQQYLAIAVH
MAB_0127c|M.abscessus_ATCC_199 ------------MATTDLHARTSSPFLELLHDQIRNEFTASQQYIAIAVY
.: * *:::*: .*: *:***:*:**.
Mb3871|M.bovis_AF2122/97 FDSEDLPQLAKHFYSQAVEERNHAMMLVQHLLDRDLRVEIPGVDTVRNQF
Rv3841|M.tuberculosis_H37Rv FDSEDLPQLAKHFYSQAVEERNHAMMLVQHLLDRDLRVEIPGVDTVRNQF
MMAR_5393|M.marinum_M FDGEELPQLAKHFYAQAVEERNHAMMLVQHLIDRDLRVEIPGIDSVRNHF
MUL_5014|M.ulcerans_Agy99 FDSEELPQLAKHFYAQAVEERNHAMMLVQHLIDRELRVEIPGIDSVRNHF
MSMEG_6422|M.smegmatis_MC2_155 FDGADLPQLAKHFYAQALEERNHAMMLVQYLLDRDVEVEIPGIDPVCNNF
Mflv_1141|M.gilvum_PYR-GCK FDGADLPQLATLFYRQSVEERNHAMMLVRYLLDRDVHAEIPAIDPVRNTF
Mvan_5666|M.vanbaalenii_PYR-1 FDSSDLPQLAKFFYRQSVEERNHAMMLVQYLIDRGVPAEIPAVDPVRNTF
MAB_0127c|M.abscessus_ATCC_199 FDDADLPQLAKRFYAQAVEERNHAMMIIRYLIDKNVAITIPGVDAVVTEF
**. :*****. ** *::********::::*:*: : **.:*.* . *
Mb3871|M.bovis_AF2122/97 DRPREALALALDQERTVTDQVGRLTAVARDEGDFLGEQFMQWFLQEQIEE
Rv3841|M.tuberculosis_H37Rv DRPREALALALDQERTVTDQVGRLTAVARDEGDFLGEQFMQWFLQEQIEE
MMAR_5393|M.marinum_M DKPRDALALALDQERAVTDQVSRLTAVARDESDFLGEQFMQWFLQEQIEE
MUL_5014|M.ulcerans_Agy99 DKPRDALALALDQERAVTDQVSRLTAVARDESDFLGEQFMQWFLQEQIEE
MSMEG_6422|M.smegmatis_MC2_155 TTPRDALALALDQERTVTEQISRLASVARDEGDHLGEQFMQWFLKEQVEE
Mflv_1141|M.gilvum_PYR-GCK DTARDALGLALDLERSVTEQISRLAATARDEGDYLGEQFMQWFLKEQVEE
Mvan_5666|M.vanbaalenii_PYR-1 ETPREAIALALDLERAVTEQISRLAGTARDEGDYLGEQFMQWFLKEQVEE
MAB_0127c|M.abscessus_ATCC_199 ADARAPIALALEQEKRVTDQVTELARAARSAGDYYGEQFMQWFLKEQVEE
.* .:.***: *: **:*: .*: .**. .*. *********:**:**
Mb3871|M.bovis_AF2122/97 VALMATLVRVADRAGANLFELENFVAREVD-VAPAASGAPHAAGGRL
Rv3841|M.tuberculosis_H37Rv VALMATLVRVADRAGANLFELENFVAREVD-VAPAASGAPHAAGGRL
MMAR_5393|M.marinum_M VALMASLVRIADRAGANLFELENFVAREVG-RAPSGAAAPRAAGGNL
MUL_5014|M.ulcerans_Agy99 VALMASLVRIADRAGANLFELENFVAREVG-RAPSGAAAPRAAGGNL
MSMEG_6422|M.smegmatis_MC2_155 VAAMTTLVRIADRAGSNLFHIEDFVAREMS-AAGADPTAPRAAGGAL
Mflv_1141|M.gilvum_PYR-GCK VATMTTLVRIAERAGADLFHLEDFVAREIS-ASAADPAAPTAAGGTL
Mvan_5666|M.vanbaalenii_PYR-1 VALMTTLVRIADRAGADLFQLEDYVAREIT-PGAADPSAPRAAGGAL
MAB_0127c|M.abscessus_ATCC_199 VALMDTLLTVAERADHDLFDLENFVARELARPVSPDPTAPTAAGGAL
** * :*: :*:**. :**.:*::****: . . ** **** *