For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGQPDTLAEARWTSERNDNRMQGLVLQLTRGGFLLLLWVFIWSVLRILRTDIYAPTGQLMANRGLNLRSI RPAKTNRRQMQYLLVTEGALAGTRITLGTQPILIGRADDSTLVLTDDYSSTRHARLSPRGSDWYVEDLGS TNGTYLDREKVTTALRVPQGTPVRIGKTVIELRS*
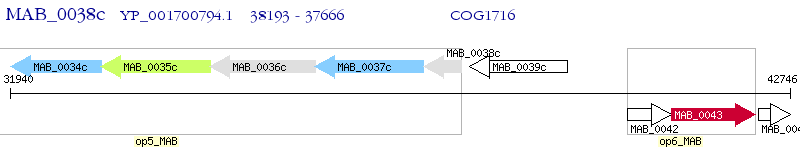
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0038c | - | - | 100% (175) | forkhead-associated protein |
| M. abscessus ATCC 19977 | MAB_2403 | - | 2e-09 | 40.00% (85) | hypothetical protein MAB_2403 |
| M. abscessus ATCC 19977 | MAB_3698 | - | 3e-05 | 35.38% (65) | ABC transporter ATP-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0019c | - | 2e-64 | 78.57% (154) | hypothetical protein Mb0019c |
| M. gilvum PYR-GCK | Mflv_0807 | - | 2e-67 | 79.87% (159) | FHA domain-containing protein |
| M. tuberculosis H37Rv | Rv0019c | - | 2e-64 | 78.57% (154) | hypothetical protein Rv0019c |
| M. leprae Br4923 | MLBr_00021 | - | 3e-62 | 74.68% (154) | hypothetical protein MLBr_00021 |
| M. marinum M | MMAR_0021 | - | 6e-65 | 77.92% (154) | hypothetical protein MMAR_0021 |
| M. avium 104 | MAV_0023 | - | 5e-52 | 76.34% (131) | FHA domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_0034 | - | 2e-66 | 81.17% (154) | FHA domain-containing protein |
| M. thermoresistible (build 8) | TH_2238 | - | 2e-66 | 81.17% (154) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0023 | - | 2e-64 | 77.27% (154) | hypothetical protein MUL_0023 |
| M. vanbaalenii PYR-1 | Mvan_0028 | - | 1e-67 | 79.87% (159) | FHA domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0019c|M.bovis_AF2122/97 ---------------------MQGLVLQLTRAGFLMLLWVFIWSVLRILK
Rv0019c|M.tuberculosis_H37Rv ---------------------MQGLVLQLTRAGFLMLLWVFIWSVLRILK
MLBr_00021|M.leprae_Br4923 ---------------------MQGLVLQLARAGFLMLLWVFIWSVLQILK
MAV_0023|M.avium_104 --------------------------------------------MLRILR
MMAR_0021|M.marinum_M ---------------------MQGLVLQLTRAGFLMLLWLFIWSVLRILR
MUL_0023|M.ulcerans_Agy99 ---------------------MQGLVLQLTRAGFLMLLWLFIWSVLRILR
Mflv_0807|M.gilvum_PYR-GCK --------MREEHDRVRRGRQMQGLVLQLTRVGFLLLLWLFIWSVLRILR
Mvan_0028|M.vanbaalenii_PYR-1 --------MREEYQRRRRGRQMQGLVLQLTRVGFLLLLWLFIWSVLRILR
MSMEG_0034|M.smegmatis_MC2_155 ---------------------MQGLVLQLTRVGFLLLLWLFIWSVLRILR
TH_2238|M.thermoresistible__bu ---------------------MQGLVLQLTRVGFLLLLWLFIWSVLRILR
MAB_0038c|M.abscessus_ATCC_199 MSGQPDTLAEARWTSERNDNRMQGLVLQLTRGGFLLLLWVFIWSVLRILR
:*:**:
Mb0019c|M.bovis_AF2122/97 TDIYAPTGAVMMRRGLALRGTLLGARQRRHAARYLVVTEGALTGARITLS
Rv0019c|M.tuberculosis_H37Rv TDIYAPTGAVMMRRGLALRGTLLGARQRRHAARYLVVTEGALTGARITLS
MLBr_00021|M.leprae_Br4923 TDIYAPTGAVMVRRSLTLRNTLLLSRQRRHAARYLMVTEGSLTGARITLS
MAV_0023|M.avium_104 TDIYAPTGAVMVRRGLALRSTLLPSRQRRHAARYLVVTEGALAGGRITLS
MMAR_0021|M.marinum_M TDIYAPTGAVMVRRGLALRGTLLPSRQRGQSARYLVVTEGALTGARITLS
MUL_0023|M.ulcerans_Agy99 TDIYAPTGAVMVRRGLALRGTLLPSRRRGQSARYLVVTEGALTGARITLS
Mflv_0807|M.gilvum_PYR-GCK TDIYAPTGAVMVRRGLALRGSLLPHRGRRTTPRQLVVTEGALAGTRIPLG
Mvan_0028|M.vanbaalenii_PYR-1 TDIYAPTGAVMVRRGLALRGSLLPNRSRRHVPRQLVVTEGALAGTRIPLG
MSMEG_0034|M.smegmatis_MC2_155 TDIYAPTGAVMVRRGLALRGSLLPNRQRRHVARQLVVTEGALAGTRITLG
TH_2238|M.thermoresistible__bu TDIYAPTGAVMMRRGLPLRGSLLPSRQRRNVARQLVVVEGALAGTRIPLG
MAB_0038c|M.abscessus_ATCC_199 TDIYAPTGQLMANRGLNLR-SIRPAKTNRRQMQYLLVTEGALAGTRITLG
******** :* .*.* ** :: : . : *:*.**:*:* **.*.
Mb0019c|M.bovis_AF2122/97 EQPVLIGRADDSTLVLTDDYASTRHARLSMRGSEWYVEDLGSTNGTYLDR
Rv0019c|M.tuberculosis_H37Rv EQPVLIGRADDSTLVLTDDYASTRHARLSMRGSEWYVEDLGSTNGTYLDR
MLBr_00021|M.leprae_Br4923 GQPVLIGRADDSTLVLTDDYASNRHARLSQRGSEWYVEDLGSTNGTYLDR
MAV_0023|M.avium_104 GQPVLIGRADDSTLVLTDDYASARHARLTQRGSEWYVEDLGSTNGTYLDR
MMAR_0021|M.marinum_M GQPVLIGRADDSTLVLTDDYASTRHARLSQRGSEWYVEDLGSTNGTYLDR
MUL_0023|M.ulcerans_Agy99 GQPVLIGRADDSTLVLTDDYASTRHARLSQRGSEWYVEDLGSTNGTYLDR
Mflv_0807|M.gilvum_PYR-GCK TQPVLIGRADDSTLVLTDDYASTRHARLSPRGPEWYVEDLGSTNGTYLDR
Mvan_0028|M.vanbaalenii_PYR-1 TQPVLIGRADDSTLVLTDDYASTRHARLSPRGPEWYVEDLGSTNGTYLDR
MSMEG_0034|M.smegmatis_MC2_155 NQPVLIGRADDSTLVLTDDYASTRHARLSPRGSEWYVEDLGSTNGTYLDR
TH_2238|M.thermoresistible__bu SQPVLIGRADDSTLVLTDDYASTRHARLSPRGSEWYVEDLGSTNGTYLDR
MAB_0038c|M.abscessus_ATCC_199 TQPILIGRADDSTLVLTDDYSSTRHARLSPRGSDWYVEDLGSTNGTYLDR
**:****************:* *****: **.:****************
Mb0019c|M.bovis_AF2122/97 AKVTTAVRVPIGTPVRIGKTAIELRP
Rv0019c|M.tuberculosis_H37Rv AKVTTAVRVPIGTPVRIGKTAIELRP
MLBr_00021|M.leprae_Br4923 AKVTTAVRVPLGTPVRIGKTAIELRP
MAV_0023|M.avium_104 AKVTTAVRVPIGTPIRIGKTAIELRP
MMAR_0021|M.marinum_M AKVTTAVRVPMGTPVRIGKTAIELRP
MUL_0023|M.ulcerans_Agy99 AKVTAAVRVPMGTPVRIGKTAIELRP
Mflv_0807|M.gilvum_PYR-GCK AKVTTAVRVPMGTPVRIGKTVIELRP
Mvan_0028|M.vanbaalenii_PYR-1 AKVTTAVRVPMGTPVRIGKTVIELRP
MSMEG_0034|M.smegmatis_MC2_155 AKVTTAVKVPIGAPVRIGKTVIELRP
TH_2238|M.thermoresistible__bu VKVTTAVRVPIGTPVRIGKTVIELRP
MAB_0038c|M.abscessus_ATCC_199 EKVTTALRVPQGTPVRIGKTVIELRS
***:*::** *:*:*****.****.